Polarized Image and Instrumental Modeling
In this tutorial, we will analyze a simulated simple polarized dataset to demonstrate Comrade's polarized imaging capabilities.
Introduction to Polarized Imaging
The EHT is a polarized interferometer. However, like all VLBI interferometers, it does not directly measure the Stokes parameters (I, Q, U, V). Instead, it measures components related to the electric field at the telescope along two directions using feeds. There are two types of feeds at telescopes: circular, which measure
These coherency matrices are the fundamental object in interferometry and what the telescope observes. For a perfect interferometer, these circular coherency matrices are related to the usual Fourier transform of the stokes parameters by
Note
In this tutorial, we stick to circular feeds but Comrade has the capabilities to model linear (XX,XY, ...) and mixed basis coherencies (e.g., RX, RY, ...).
In reality, the measure coherencies are corrupted by both the atmosphere and the telescope itself. In Comrade we use the RIME formalism [1] to represent these corruptions, namely our measured coherency matrices
where
Comrade is highly flexible with how the Jones matrices are formed and provides several convenience functions that parameterize standard Jones matrices. These matrices include:
JonesGwhich builds the set of complex gain Jones matrices
JonesDwhich builds the set of complex d-terms Jones matrices
JonesRis the ideal telescope response. This transformation is special and combines two things using the decomposition . The first, , is the transformation from some reference basis to the observed coherency basis (this allows for mixed basis measurements). The second is the feed rotation, , that transforms from some reference axis to the axis of the telescope as the source moves in the sky. The feed rotation matrix Ffor circular feeds in terms of the per station feed rotation angleis
In the rest of the tutorial, we are going to solve for all of these instrument model terms in while re-creating the polarized image from the first EHT results on M87.
Load the Data
To get started we will load Comrade
using ComradeLoad the Data
using Pyehtim CondaPkg Found dependencies: /home/runner/.julia/packages/DimensionalData/hv9KC/CondaPkg.toml
CondaPkg Found dependencies: /home/runner/.julia/packages/CondaPkg/0UqYV/CondaPkg.toml
CondaPkg Found dependencies: /home/runner/.julia/packages/PythonCall/83z4q/CondaPkg.toml
CondaPkg Found dependencies: /home/runner/.julia/packages/Pyehtim/bQtHC/CondaPkg.toml
CondaPkg Resolving changes
+ ehtim (pip)
+ libstdcxx
+ libstdcxx-ng
+ numpy
+ numpy (pip)
+ openssl
+ pandas
+ python
+ setuptools (pip)
+ uv
+ xarray
CondaPkg Initialising pixi
│ /home/runner/.julia/artifacts/cefba4912c2b400756d043a2563ef77a0088866b/bin/pixi
│ init
│ --format pixi
└ /home/runner/work/Comrade.jl/Comrade.jl/examples/intermediate/PolarizedImaging/.CondaPkg
✔ Created /home/runner/work/Comrade.jl/Comrade.jl/examples/intermediate/PolarizedImaging/.CondaPkg/pixi.toml
CondaPkg Wrote /home/runner/work/Comrade.jl/Comrade.jl/examples/intermediate/PolarizedImaging/.CondaPkg/pixi.toml
│ [dependencies]
│ openssl = ">=3, <3.6, >=3, <3.6"
│ libstdcxx = ">=3.4,<15.0"
│ uv = ">=0.4"
│ libstdcxx-ng = ">=3.4,<15.0"
│ pandas = "<2"
│ xarray = "*"
│ numpy = ">=1.24, <2.0"
│
│ [dependencies.python]
│ channel = "conda-forge"
│ build = "*cp*"
│ version = ">=3.10,!=3.14.0,!=3.14.1,<4, >=3.6,<=3.12"
│
│ [project]
│ name = ".CondaPkg"
│ platforms = ["linux-64"]
│ channels = ["conda-forge"]
│ channel-priority = "strict"
│ description = "automatically generated by CondaPkg.jl"
│
│ [pypi-dependencies]
│ ehtim = ">=1.2.10, <2.0"
│ numpy = ">=1.24, <2.0"
└ setuptools = "*"
CondaPkg Installing packages
│ /home/runner/.julia/artifacts/cefba4912c2b400756d043a2563ef77a0088866b/bin/pixi
│ install
└ --manifest-path /home/runner/work/Comrade.jl/Comrade.jl/examples/intermediate/PolarizedImaging/.CondaPkg/pixi.toml
✔ The default environment has been installed.
/home/runner/work/Comrade.jl/Comrade.jl/examples/intermediate/PolarizedImaging/.CondaPkg/.pixi/envs/default/lib/python3.11/site-packages/ehtim/__init__.py:58: UserWarning: pkg_resources is deprecated as an API. See https://setuptools.pypa.io/en/latest/pkg_resources.html. The pkg_resources package is slated for removal as early as 2025-11-30. Refrain from using this package or pin to Setuptools<81.
import pkg_resourcesFor reproducibility we use a stable random number genreator
using StableRNGs
rng = StableRNG(48)StableRNGs.LehmerRNG(state=0x00000000000000000000000000000061)Now we will load some synthetic polarized data.
fname = Base.download(
"https://de.cyverse.org/anon-files/iplant/home/shared/commons_repo/curated/EHTC_M87pol2017_Nov2023/hops_data/April11/SR2_M87_2017_101_lo_hops_ALMArot.uvfits",
joinpath(__DIR, "m87polarized.uvfits")
)
obs = Pyehtim.load_uvfits_and_array(
fname,
joinpath(__DIR, "..", "..", "Data", "array.txt"), polrep = "circ"
)Python: <ehtim.obsdata.Obsdata object at 0x7fba8f723310>Notice that, unlike other non-polarized tutorials, we need to include a second argument. This is the array file of the observation and is required to determine the feed rotation of the array.
Now we scan average the data since the data to boost the SNR and reduce the total data volume.
obs = scan_average(obs).add_fractional_noise(0.01).flag_uvdist(uv_min = 0.1e9)Python: <ehtim.obsdata.Obsdata object at 0x7fba8e58c610>Now we extract our observed/corrupted coherency matrices.
dvis = extract_table(obs, Coherencies())EHTObservationTable{Comrade.EHTCoherencyDatum{Float64}}
source: M87
mjd: 57854
bandwidth: 1.856e9
sites: [:AA, :AP, :AZ, :JC, :LM, :PV, :SM]
nsamples: 189##Building the Model/Posterior
To build the model, we first break it down into two parts:
The image or sky model. In Comrade, all polarized image models are written in terms of the Stokes parameters. In this tutorial, we will use a polarized image model based on Pesce (2021)[2], and parameterizes each pixel in terms of the
Poincare sphere. This parameterization ensures that we have all the physical properties of stokes parameters. Note that we also have a parameterization in terms of hyperbolic trig functionsVLBISkyModels.PolExp2MapThe instrument model. The instrument model specifies the model that describes the impact of instrumental and atmospheric effects. We will be using the
decomposition we described above. However, to parameterize the R/L complex gains, we will be using a gain product and ratio decomposition. The reason for this decomposition is that in realistic measurements, the gain ratios and products have different temporal characteristics. Namely, many of the EHT observations tend to demonstrate constant R/L gain ratios across an nights observations, compared to the gain products, which vary every scan. Additionally, the gain ratios tend to be smaller (i.e., closer to unity) than the gain products. Using this apriori knowledge, we can build this into our model and reduce the total number of parameters we need to model.
First we specify our sky model. As always Comrade requires this to be a two argument function where the first argument is typically a NamedTuple of parameters we will fit and the second are additional metadata required to build the model.
function sky(θ, metadata)
(; σs, as) = θ
(; mimg, ftot) = metadata
# Build the stokes I model
δs = ntuple(Val(4)) do i
σs[i] * as[i].params
end
# Convert hyperbolic polarization basis to Stokes basis
pmap = VLBISkyModels.PolExp2Map!(δs..., axisdims(mimg))
# We now add a mean image. Namely, we assume that `pmap` are multiplicative fluctuations
# about some mean image `mimg`. We also compute the total flux of the Stokes I image
# for normalization purposes below.
ft = zero(eltype(mimg))
for i in eachindex(pmap, mimg)
pmap[i] *= mimg[i]
ft += pmap[i].I
end
pmap .= ftot .* pmap ./ ft
m = ContinuousImage(pmap, BSplinePulse{3}())
x, y = centroid(pmap)
return shifted(m, -x, -y)
endsky (generic function with 1 method)Now, we define the model metadata required to build the model. We specify our image grid and cache model needed to define the polarimetric image model. Our image will be a 10x10 raster with a 60μas FOV.
using Distributions
using VLBIImagePriors
fovx = μas2rad(200.0)
fovy = μas2rad(200.0)
nx = ny = 48
grid = imagepixels(fovx, fovy, nx, ny)
fwhmfac = 2 * sqrt(2 * log(2))
mpr = modify(Gaussian(), Stretch(μas2rad(50.0) ./ fwhmfac))
mimg = intensitymap(mpr, grid)┌ 48×48 IntensityMap{Float64, 2} ┐
├────────────────────────────────┴─────────────────────────────────────── dims ┐
↓ X Sampled{Float64} LinRange{Float64}(-4.747133960864206e-10, 4.747133960864206e-10, 48) ForwardOrdered Regular Points,
→ Y Sampled{Float64} LinRange{Float64}(-4.747133960864206e-10, 4.747133960864206e-10, 48) ForwardOrdered Regular Points
└──────────────────────────────────────────────────────────────────────────────┘
↓ → -4.74713e-10 -4.54513e-10 -4.34312e-10 -4.14112e-10 -3.93911e-10 -3.73711e-10 -3.5351e-10 -3.33309e-10 -3.13109e-10 -2.92908e-10 -2.72708e-10 -2.52507e-10 -2.32307e-10 -2.12106e-10 -1.91905e-10 -1.71705e-10 -1.51504e-10 -1.31304e-10 -1.11103e-10 -9.09026e-11 -7.0702e-11 -5.05014e-11 -3.03009e-11 -1.01003e-11 1.01003e-11 3.03009e-11 5.05014e-11 7.0702e-11 9.09026e-11 1.11103e-10 1.31304e-10 1.51504e-10 1.71705e-10 1.91905e-10 2.12106e-10 2.32307e-10 2.52507e-10 2.72708e-10 2.92908e-10 3.13109e-10 3.33309e-10 3.5351e-10 3.73711e-10 3.93911e-10 4.14112e-10 4.34312e-10 4.54513e-10 4.74713e-10
-4.74713e-10 0.0 0.0 0.0 0.0 9.76922e-11 2.03055e-10 4.0611e-10 7.81538e-10 1.44721e-9 2.57864e-9 4.42105e-9 7.29349e-9 1.15777e-8 1.76842e-8 2.59911e-8 3.67569e-8 5.00184e-8 6.54933e-8 8.25164e-8 1.00037e-7 1.16696e-7 1.30987e-7 1.41473e-7 1.47028e-7 1.47028e-7 1.41473e-7 1.30987e-7 1.16696e-7 1.00037e-7 8.25164e-8 6.54933e-8 5.00184e-8 3.67569e-8 2.59911e-8 1.76842e-8 1.15777e-8 7.29349e-9 4.42105e-9 2.57864e-9 1.44721e-9 7.81538e-10 4.0611e-10 2.03055e-10 9.76922e-11 0.0 0.0 0.0 0.0
-4.54513e-10 0.0 0.0 0.0 1.09656e-10 2.3687e-10 4.92338e-10 9.84676e-10 1.89496e-9 3.50899e-9 6.2523e-9 1.07195e-8 1.76842e-8 2.80719e-8 4.2878e-8 6.30193e-8 8.91227e-8 1.21277e-7 1.58799e-7 2.00074e-7 2.42555e-7 2.82947e-7 3.17597e-7 3.43024e-7 3.56491e-7 3.56491e-7 3.43024e-7 3.17597e-7 2.82947e-7 2.42555e-7 2.00074e-7 1.58799e-7 1.21277e-7 8.91227e-8 6.30193e-8 4.2878e-8 2.80719e-8 1.76842e-8 1.07195e-8 6.2523e-9 3.50899e-9 1.89496e-9 9.84676e-10 4.92338e-10 2.3687e-10 1.09656e-10 0.0 0.0 0.0
-4.34312e-10 0.0 0.0 1.13961e-10 2.55833e-10 5.52631e-10 1.14865e-9 2.29731e-9 4.42105e-9 8.18667e-9 1.4587e-8 2.50092e-8 4.12582e-8 6.54933e-8 1.00037e-7 1.47028e-7 2.07928e-7 2.82947e-7 3.70486e-7 4.66784e-7 5.65894e-7 6.60132e-7 7.40973e-7 8.00295e-7 8.31714e-7 8.31714e-7 8.00295e-7 7.40973e-7 6.60132e-7 5.65894e-7 4.66784e-7 3.70486e-7 2.82947e-7 2.07928e-7 1.47028e-7 1.00037e-7 6.54933e-8 4.12582e-8 2.50092e-8 1.4587e-8 8.18667e-9 4.42105e-9 2.29731e-9 1.14865e-9 5.52631e-10 2.55833e-10 1.13961e-10 0.0 0.0
-4.14112e-10 0.0 1.09656e-10 2.55833e-10 5.74327e-10 1.24061e-9 2.57864e-9 5.15728e-9 9.92491e-9 1.83784e-8 3.27467e-8 5.61438e-8 9.26216e-8 1.47028e-7 2.24575e-7 3.30066e-7 4.66784e-7 6.35194e-7 8.31714e-7 1.04789e-6 1.27039e-6 1.48195e-6 1.66343e-6 1.7966e-6 1.86713e-6 1.86713e-6 1.7966e-6 1.66343e-6 1.48195e-6 1.27039e-6 1.04789e-6 8.31714e-7 6.35194e-7 4.66784e-7 3.30066e-7 2.24575e-7 1.47028e-7 9.26216e-8 5.61438e-8 3.27467e-8 1.83784e-8 9.92491e-9 5.15728e-9 2.57864e-9 1.24061e-9 5.74327e-10 2.55833e-10 1.09656e-10 0.0
-3.93911e-10 9.76922e-11 2.3687e-10 5.52631e-10 1.24061e-9 2.67987e-9 5.57017e-9 1.11403e-8 2.1439e-8 3.96996e-8 7.07367e-8 1.21277e-7 2.00074e-7 3.17597e-7 4.85109e-7 7.12982e-7 1.00831e-6 1.3721e-6 1.7966e-6 2.26358e-6 2.74419e-6 3.20118e-6 3.5932e-6 3.88087e-6 4.03323e-6 4.03323e-6 3.88087e-6 3.5932e-6 3.20118e-6 2.74419e-6 2.26358e-6 1.7966e-6 1.3721e-6 1.00831e-6 7.12982e-7 4.85109e-7 3.17597e-7 2.00074e-7 1.21277e-7 7.07367e-8 3.96996e-8 2.1439e-8 1.11403e-8 5.57017e-9 2.67987e-9 1.24061e-9 5.52631e-10 2.3687e-10 9.76922e-11
-3.73711e-10 2.03055e-10 4.92338e-10 1.14865e-9 2.57864e-9 5.57017e-9 1.15777e-8 2.31554e-8 4.45613e-8 8.25164e-8 1.47028e-7 2.52077e-7 4.15857e-7 6.60132e-7 1.00831e-6 1.48195e-6 2.09579e-6 2.85193e-6 3.73427e-6 4.70488e-6 5.70385e-6 6.65371e-6 7.46854e-6 8.06647e-6 8.38315e-6 8.38315e-6 8.06647e-6 7.46854e-6 6.65371e-6 5.70385e-6 4.70488e-6 3.73427e-6 2.85193e-6 2.09579e-6 1.48195e-6 1.00831e-6 6.60132e-7 4.15857e-7 2.52077e-7 1.47028e-7 8.25164e-8 4.45613e-8 2.31554e-8 1.15777e-8 5.57017e-9 2.57864e-9 1.14865e-9 4.92338e-10 2.03055e-10
-3.5351e-10 4.0611e-10 9.84676e-10 2.29731e-9 5.15728e-9 1.11403e-8 2.31554e-8 4.63108e-8 8.91227e-8 1.65033e-7 2.94055e-7 5.04154e-7 8.31714e-7 1.32026e-6 2.01662e-6 2.96389e-6 4.19157e-6 5.70385e-6 7.46854e-6 9.40977e-6 1.14077e-5 1.33074e-5 1.49371e-5 1.61329e-5 1.67663e-5 1.67663e-5 1.61329e-5 1.49371e-5 1.33074e-5 1.14077e-5 9.40977e-6 7.46854e-6 5.70385e-6 4.19157e-6 2.96389e-6 2.01662e-6 1.32026e-6 8.31714e-7 5.04154e-7 2.94055e-7 1.65033e-7 8.91227e-8 4.63108e-8 2.31554e-8 1.11403e-8 5.15728e-9 2.29731e-9 9.84676e-10 4.0611e-10
-3.33309e-10 7.81538e-10 1.89496e-9 4.42105e-9 9.92491e-9 2.1439e-8 4.45613e-8 8.91227e-8 1.71512e-7 3.17597e-7 5.65894e-7 9.70218e-7 1.60059e-6 2.54078e-6 3.88087e-6 5.70385e-6 8.06647e-6 1.09768e-5 1.43728e-5 1.81086e-5 2.19535e-5 2.56094e-5 2.87456e-5 3.1047e-5 3.22659e-5 3.22659e-5 3.1047e-5 2.87456e-5 2.56094e-5 2.19535e-5 1.81086e-5 1.43728e-5 1.09768e-5 8.06647e-6 5.70385e-6 3.88087e-6 2.54078e-6 1.60059e-6 9.70218e-7 5.65894e-7 3.17597e-7 1.71512e-7 8.91227e-8 4.45613e-8 2.1439e-8 9.92491e-9 4.42105e-9 1.89496e-9 7.81538e-10
-3.13109e-10 1.44721e-9 3.50899e-9 8.18667e-9 1.83784e-8 3.96996e-8 8.25164e-8 1.65033e-7 3.17597e-7 5.8811e-7 1.04789e-6 1.7966e-6 2.96389e-6 4.70488e-6 7.1864e-6 1.05621e-5 1.49371e-5 2.03262e-5 2.66148e-5 3.35326e-5 4.06524e-5 4.74222e-5 5.32297e-5 5.74912e-5 5.97483e-5 5.97483e-5 5.74912e-5 5.32297e-5 4.74222e-5 4.06524e-5 3.35326e-5 2.66148e-5 2.03262e-5 1.49371e-5 1.05621e-5 7.1864e-6 4.70488e-6 2.96389e-6 1.7966e-6 1.04789e-6 5.8811e-7 3.17597e-7 1.65033e-7 8.25164e-8 3.96996e-8 1.83784e-8 8.18667e-9 3.50899e-9 1.44721e-9
-2.92908e-10 2.57864e-9 6.2523e-9 1.4587e-8 3.27467e-8 7.07367e-8 1.47028e-7 2.94055e-7 5.65894e-7 1.04789e-6 1.86713e-6 3.20118e-6 5.28105e-6 8.38315e-6 1.28047e-5 1.88195e-5 2.66148e-5 3.62172e-5 4.74222e-5 5.97483e-5 7.24344e-5 8.44968e-5 9.48445e-5 0.000102438 0.000106459 0.000106459 0.000102438 9.48445e-5 8.44968e-5 7.24344e-5 5.97483e-5 4.74222e-5 3.62172e-5 2.66148e-5 1.88195e-5 1.28047e-5 8.38315e-6 5.28105e-6 3.20118e-6 1.86713e-6 1.04789e-6 5.65894e-7 2.94055e-7 1.47028e-7 7.07367e-8 3.27467e-8 1.4587e-8 6.2523e-9 2.57864e-9
-2.72708e-10 4.42105e-9 1.07195e-8 2.50092e-8 5.61438e-8 1.21277e-7 2.52077e-7 5.04154e-7 9.70218e-7 1.7966e-6 3.20118e-6 5.48838e-6 9.0543e-6 1.43728e-5 2.19535e-5 3.22659e-5 4.56308e-5 6.2094e-5 8.13049e-5 0.000102438 0.000124188 0.000144869 0.00016261 0.000175628 0.000182523 0.000182523 0.000175628 0.00016261 0.000144869 0.000124188 0.000102438 8.13049e-5 6.2094e-5 4.56308e-5 3.22659e-5 2.19535e-5 1.43728e-5 9.0543e-6 5.48838e-6 3.20118e-6 1.7966e-6 9.70218e-7 5.04154e-7 2.52077e-7 1.21277e-7 5.61438e-8 2.50092e-8 1.07195e-8 4.42105e-9
-2.52507e-10 7.29349e-9 1.76842e-8 4.12582e-8 9.26216e-8 2.00074e-7 4.15857e-7 8.31714e-7 1.60059e-6 2.96389e-6 5.28105e-6 9.0543e-6 1.49371e-5 2.37111e-5 3.62172e-5 5.32297e-5 7.52781e-5 0.000102438 0.00013413 0.000168994 0.000204875 0.000238993 0.000268261 0.000289738 0.000301113 0.000301113 0.000289738 0.000268261 0.000238993 0.000204875 0.000168994 0.00013413 0.000102438 7.52781e-5 5.32297e-5 3.62172e-5 2.37111e-5 1.49371e-5 9.0543e-6 5.28105e-6 2.96389e-6 1.60059e-6 8.31714e-7 4.15857e-7 2.00074e-7 9.26216e-8 4.12582e-8 1.76842e-8 7.29349e-9
-2.32307e-10 1.15777e-8 2.80719e-8 6.54933e-8 1.47028e-7 3.17597e-7 6.60132e-7 1.32026e-6 2.54078e-6 4.70488e-6 8.38315e-6 1.43728e-5 2.37111e-5 3.76391e-5 5.74912e-5 8.44968e-5 0.000119497 0.00016261 0.000212919 0.000268261 0.00032522 0.000379378 0.000425837 0.00045993 0.000477986 0.000477986 0.00045993 0.000425837 0.000379378 0.00032522 0.000268261 0.000212919 0.00016261 0.000119497 8.44968e-5 5.74912e-5 3.76391e-5 2.37111e-5 1.43728e-5 8.38315e-6 4.70488e-6 2.54078e-6 1.32026e-6 6.60132e-7 3.17597e-7 1.47028e-7 6.54933e-8 2.80719e-8 1.15777e-8
-2.12106e-10 1.76842e-8 4.2878e-8 1.00037e-7 2.24575e-7 4.85109e-7 1.00831e-6 2.01662e-6 3.88087e-6 7.1864e-6 1.28047e-5 2.19535e-5 3.62172e-5 5.74912e-5 8.78141e-5 0.000129063 0.000182523 0.000248376 0.00032522 0.000409751 0.000496752 0.000579475 0.000650439 0.000702513 0.000730093 0.000730093 0.000702513 0.000650439 0.000579475 0.000496752 0.000409751 0.00032522 0.000248376 0.000182523 0.000129063 8.78141e-5 5.74912e-5 3.62172e-5 2.19535e-5 1.28047e-5 7.1864e-6 3.88087e-6 2.01662e-6 1.00831e-6 4.85109e-7 2.24575e-7 1.00037e-7 4.2878e-8 1.76842e-8
-1.91905e-10 2.59911e-8 6.30193e-8 1.47028e-7 3.30066e-7 7.12982e-7 1.48195e-6 2.96389e-6 5.70385e-6 1.05621e-5 1.88195e-5 3.22659e-5 5.32297e-5 8.44968e-5 0.000129063 0.000189689 0.000268261 0.000365047 0.000477986 0.000602225 0.000730093 0.000851675 0.000955973 0.00103251 0.00107304 0.00107304 0.00103251 0.000955973 0.000851675 0.000730093 0.000602225 0.000477986 0.000365047 0.000268261 0.000189689 0.000129063 8.44968e-5 5.32297e-5 3.22659e-5 1.88195e-5 1.05621e-5 5.70385e-6 2.96389e-6 1.48195e-6 7.12982e-7 3.30066e-7 1.47028e-7 6.30193e-8 2.59911e-8
-1.71705e-10 3.67569e-8 8.91227e-8 2.07928e-7 4.66784e-7 1.00831e-6 2.09579e-6 4.19157e-6 8.06647e-6 1.49371e-5 2.66148e-5 4.56308e-5 7.52781e-5 0.000119497 0.000182523 0.000268261 0.000379378 0.000516254 0.000675975 0.000851675 0.00103251 0.00120445 0.00135195 0.00146019 0.00151751 0.00151751 0.00146019 0.00135195 0.00120445 0.00103251 0.000851675 0.000675975 0.000516254 0.000379378 0.000268261 0.000182523 0.000119497 7.52781e-5 4.56308e-5 2.66148e-5 1.49371e-5 8.06647e-6 4.19157e-6 2.09579e-6 1.00831e-6 4.66784e-7 2.07928e-7 8.91227e-8 3.67569e-8
-1.51504e-10 5.00184e-8 1.21277e-7 2.82947e-7 6.35194e-7 1.3721e-6 2.85193e-6 5.70385e-6 1.09768e-5 2.03262e-5 3.62172e-5 6.2094e-5 0.000102438 0.00016261 0.000248376 0.000365047 0.000516254 0.000702513 0.00091986 0.00115895 0.00140503 0.001639 0.00183972 0.00198701 0.00206502 0.00206502 0.00198701 0.00183972 0.001639 0.00140503 0.00115895 0.00091986 0.000702513 0.000516254 0.000365047 0.000248376 0.00016261 0.000102438 6.2094e-5 3.62172e-5 2.03262e-5 1.09768e-5 5.70385e-6 2.85193e-6 1.3721e-6 6.35194e-7 2.82947e-7 1.21277e-7 5.00184e-8
-1.31304e-10 6.54933e-8 1.58799e-7 3.70486e-7 8.31714e-7 1.7966e-6 3.73427e-6 7.46854e-6 1.43728e-5 2.66148e-5 4.74222e-5 8.13049e-5 0.00013413 0.000212919 0.00032522 0.000477986 0.000675975 0.00091986 0.00120445 0.00151751 0.00183972 0.00214609 0.0024089 0.00260176 0.0027039 0.0027039 0.00260176 0.0024089 0.00214609 0.00183972 0.00151751 0.00120445 0.00091986 0.000675975 0.000477986 0.00032522 0.000212919 0.00013413 8.13049e-5 4.74222e-5 2.66148e-5 1.43728e-5 7.46854e-6 3.73427e-6 1.7966e-6 8.31714e-7 3.70486e-7 1.58799e-7 6.54933e-8
-1.11103e-10 8.25164e-8 2.00074e-7 4.66784e-7 1.04789e-6 2.26358e-6 4.70488e-6 9.40977e-6 1.81086e-5 3.35326e-5 5.97483e-5 0.000102438 0.000168994 0.000268261 0.000409751 0.000602225 0.000851675 0.00115895 0.00151751 0.00191195 0.0023179 0.0027039 0.00303502 0.00327801 0.0034067 0.0034067 0.00327801 0.00303502 0.0027039 0.0023179 0.00191195 0.00151751 0.00115895 0.000851675 0.000602225 0.000409751 0.000268261 0.000168994 0.000102438 5.97483e-5 3.35326e-5 1.81086e-5 9.40977e-6 4.70488e-6 2.26358e-6 1.04789e-6 4.66784e-7 2.00074e-7 8.25164e-8
-9.09026e-11 1.00037e-7 2.42555e-7 5.65894e-7 1.27039e-6 2.74419e-6 5.70385e-6 1.14077e-5 2.19535e-5 4.06524e-5 7.24344e-5 0.000124188 0.000204875 0.00032522 0.000496752 0.000730093 0.00103251 0.00140503 0.00183972 0.0023179 0.00281005 0.00327801 0.00367944 0.00397401 0.00413003 0.00413003 0.00397401 0.00367944 0.00327801 0.00281005 0.0023179 0.00183972 0.00140503 0.00103251 0.000730093 0.000496752 0.00032522 0.000204875 0.000124188 7.24344e-5 4.06524e-5 2.19535e-5 1.14077e-5 5.70385e-6 2.74419e-6 1.27039e-6 5.65894e-7 2.42555e-7 1.00037e-7
-7.0702e-11 1.16696e-7 2.82947e-7 6.60132e-7 1.48195e-6 3.20118e-6 6.65371e-6 1.33074e-5 2.56094e-5 4.74222e-5 8.44968e-5 0.000144869 0.000238993 0.000379378 0.000579475 0.000851675 0.00120445 0.001639 0.00214609 0.0027039 0.00327801 0.00382389 0.00429217 0.0046358 0.0048178 0.0048178 0.0046358 0.00429217 0.00382389 0.00327801 0.0027039 0.00214609 0.001639 0.00120445 0.000851675 0.000579475 0.000379378 0.000238993 0.000144869 8.44968e-5 4.74222e-5 2.56094e-5 1.33074e-5 6.65371e-6 3.20118e-6 1.48195e-6 6.60132e-7 2.82947e-7 1.16696e-7
-5.05014e-11 1.30987e-7 3.17597e-7 7.40973e-7 1.66343e-6 3.5932e-6 7.46854e-6 1.49371e-5 2.87456e-5 5.32297e-5 9.48445e-5 0.00016261 0.000268261 0.000425837 0.000650439 0.000955973 0.00135195 0.00183972 0.0024089 0.00303502 0.00367944 0.00429217 0.0048178 0.00520351 0.0054078 0.0054078 0.00520351 0.0048178 0.00429217 0.00367944 0.00303502 0.0024089 0.00183972 0.00135195 0.000955973 0.000650439 0.000425837 0.000268261 0.00016261 9.48445e-5 5.32297e-5 2.87456e-5 1.49371e-5 7.46854e-6 3.5932e-6 1.66343e-6 7.40973e-7 3.17597e-7 1.30987e-7
-3.03009e-11 1.41473e-7 3.43024e-7 8.00295e-7 1.7966e-6 3.88087e-6 8.06647e-6 1.61329e-5 3.1047e-5 5.74912e-5 0.000102438 0.000175628 0.000289738 0.00045993 0.000702513 0.00103251 0.00146019 0.00198701 0.00260176 0.00327801 0.00397401 0.0046358 0.00520351 0.0056201 0.00584074 0.00584074 0.0056201 0.00520351 0.0046358 0.00397401 0.00327801 0.00260176 0.00198701 0.00146019 0.00103251 0.000702513 0.00045993 0.000289738 0.000175628 0.000102438 5.74912e-5 3.1047e-5 1.61329e-5 8.06647e-6 3.88087e-6 1.7966e-6 8.00295e-7 3.43024e-7 1.41473e-7
-1.01003e-11 1.47028e-7 3.56491e-7 8.31714e-7 1.86713e-6 4.03323e-6 8.38315e-6 1.67663e-5 3.22659e-5 5.97483e-5 0.000106459 0.000182523 0.000301113 0.000477986 0.000730093 0.00107304 0.00151751 0.00206502 0.0027039 0.0034067 0.00413003 0.0048178 0.0054078 0.00584074 0.00607005 0.00607005 0.00584074 0.0054078 0.0048178 0.00413003 0.0034067 0.0027039 0.00206502 0.00151751 0.00107304 0.000730093 0.000477986 0.000301113 0.000182523 0.000106459 5.97483e-5 3.22659e-5 1.67663e-5 8.38315e-6 4.03323e-6 1.86713e-6 8.31714e-7 3.56491e-7 1.47028e-7
1.01003e-11 1.47028e-7 3.56491e-7 8.31714e-7 1.86713e-6 4.03323e-6 8.38315e-6 1.67663e-5 3.22659e-5 5.97483e-5 0.000106459 0.000182523 0.000301113 0.000477986 0.000730093 0.00107304 0.00151751 0.00206502 0.0027039 0.0034067 0.00413003 0.0048178 0.0054078 0.00584074 0.00607005 0.00607005 0.00584074 0.0054078 0.0048178 0.00413003 0.0034067 0.0027039 0.00206502 0.00151751 0.00107304 0.000730093 0.000477986 0.000301113 0.000182523 0.000106459 5.97483e-5 3.22659e-5 1.67663e-5 8.38315e-6 4.03323e-6 1.86713e-6 8.31714e-7 3.56491e-7 1.47028e-7
3.03009e-11 1.41473e-7 3.43024e-7 8.00295e-7 1.7966e-6 3.88087e-6 8.06647e-6 1.61329e-5 3.1047e-5 5.74912e-5 0.000102438 0.000175628 0.000289738 0.00045993 0.000702513 0.00103251 0.00146019 0.00198701 0.00260176 0.00327801 0.00397401 0.0046358 0.00520351 0.0056201 0.00584074 0.00584074 0.0056201 0.00520351 0.0046358 0.00397401 0.00327801 0.00260176 0.00198701 0.00146019 0.00103251 0.000702513 0.00045993 0.000289738 0.000175628 0.000102438 5.74912e-5 3.1047e-5 1.61329e-5 8.06647e-6 3.88087e-6 1.7966e-6 8.00295e-7 3.43024e-7 1.41473e-7
5.05014e-11 1.30987e-7 3.17597e-7 7.40973e-7 1.66343e-6 3.5932e-6 7.46854e-6 1.49371e-5 2.87456e-5 5.32297e-5 9.48445e-5 0.00016261 0.000268261 0.000425837 0.000650439 0.000955973 0.00135195 0.00183972 0.0024089 0.00303502 0.00367944 0.00429217 0.0048178 0.00520351 0.0054078 0.0054078 0.00520351 0.0048178 0.00429217 0.00367944 0.00303502 0.0024089 0.00183972 0.00135195 0.000955973 0.000650439 0.000425837 0.000268261 0.00016261 9.48445e-5 5.32297e-5 2.87456e-5 1.49371e-5 7.46854e-6 3.5932e-6 1.66343e-6 7.40973e-7 3.17597e-7 1.30987e-7
7.0702e-11 1.16696e-7 2.82947e-7 6.60132e-7 1.48195e-6 3.20118e-6 6.65371e-6 1.33074e-5 2.56094e-5 4.74222e-5 8.44968e-5 0.000144869 0.000238993 0.000379378 0.000579475 0.000851675 0.00120445 0.001639 0.00214609 0.0027039 0.00327801 0.00382389 0.00429217 0.0046358 0.0048178 0.0048178 0.0046358 0.00429217 0.00382389 0.00327801 0.0027039 0.00214609 0.001639 0.00120445 0.000851675 0.000579475 0.000379378 0.000238993 0.000144869 8.44968e-5 4.74222e-5 2.56094e-5 1.33074e-5 6.65371e-6 3.20118e-6 1.48195e-6 6.60132e-7 2.82947e-7 1.16696e-7
9.09026e-11 1.00037e-7 2.42555e-7 5.65894e-7 1.27039e-6 2.74419e-6 5.70385e-6 1.14077e-5 2.19535e-5 4.06524e-5 7.24344e-5 0.000124188 0.000204875 0.00032522 0.000496752 0.000730093 0.00103251 0.00140503 0.00183972 0.0023179 0.00281005 0.00327801 0.00367944 0.00397401 0.00413003 0.00413003 0.00397401 0.00367944 0.00327801 0.00281005 0.0023179 0.00183972 0.00140503 0.00103251 0.000730093 0.000496752 0.00032522 0.000204875 0.000124188 7.24344e-5 4.06524e-5 2.19535e-5 1.14077e-5 5.70385e-6 2.74419e-6 1.27039e-6 5.65894e-7 2.42555e-7 1.00037e-7
1.11103e-10 8.25164e-8 2.00074e-7 4.66784e-7 1.04789e-6 2.26358e-6 4.70488e-6 9.40977e-6 1.81086e-5 3.35326e-5 5.97483e-5 0.000102438 0.000168994 0.000268261 0.000409751 0.000602225 0.000851675 0.00115895 0.00151751 0.00191195 0.0023179 0.0027039 0.00303502 0.00327801 0.0034067 0.0034067 0.00327801 0.00303502 0.0027039 0.0023179 0.00191195 0.00151751 0.00115895 0.000851675 0.000602225 0.000409751 0.000268261 0.000168994 0.000102438 5.97483e-5 3.35326e-5 1.81086e-5 9.40977e-6 4.70488e-6 2.26358e-6 1.04789e-6 4.66784e-7 2.00074e-7 8.25164e-8
1.31304e-10 6.54933e-8 1.58799e-7 3.70486e-7 8.31714e-7 1.7966e-6 3.73427e-6 7.46854e-6 1.43728e-5 2.66148e-5 4.74222e-5 8.13049e-5 0.00013413 0.000212919 0.00032522 0.000477986 0.000675975 0.00091986 0.00120445 0.00151751 0.00183972 0.00214609 0.0024089 0.00260176 0.0027039 0.0027039 0.00260176 0.0024089 0.00214609 0.00183972 0.00151751 0.00120445 0.00091986 0.000675975 0.000477986 0.00032522 0.000212919 0.00013413 8.13049e-5 4.74222e-5 2.66148e-5 1.43728e-5 7.46854e-6 3.73427e-6 1.7966e-6 8.31714e-7 3.70486e-7 1.58799e-7 6.54933e-8
1.51504e-10 5.00184e-8 1.21277e-7 2.82947e-7 6.35194e-7 1.3721e-6 2.85193e-6 5.70385e-6 1.09768e-5 2.03262e-5 3.62172e-5 6.2094e-5 0.000102438 0.00016261 0.000248376 0.000365047 0.000516254 0.000702513 0.00091986 0.00115895 0.00140503 0.001639 0.00183972 0.00198701 0.00206502 0.00206502 0.00198701 0.00183972 0.001639 0.00140503 0.00115895 0.00091986 0.000702513 0.000516254 0.000365047 0.000248376 0.00016261 0.000102438 6.2094e-5 3.62172e-5 2.03262e-5 1.09768e-5 5.70385e-6 2.85193e-6 1.3721e-6 6.35194e-7 2.82947e-7 1.21277e-7 5.00184e-8
1.71705e-10 3.67569e-8 8.91227e-8 2.07928e-7 4.66784e-7 1.00831e-6 2.09579e-6 4.19157e-6 8.06647e-6 1.49371e-5 2.66148e-5 4.56308e-5 7.52781e-5 0.000119497 0.000182523 0.000268261 0.000379378 0.000516254 0.000675975 0.000851675 0.00103251 0.00120445 0.00135195 0.00146019 0.00151751 0.00151751 0.00146019 0.00135195 0.00120445 0.00103251 0.000851675 0.000675975 0.000516254 0.000379378 0.000268261 0.000182523 0.000119497 7.52781e-5 4.56308e-5 2.66148e-5 1.49371e-5 8.06647e-6 4.19157e-6 2.09579e-6 1.00831e-6 4.66784e-7 2.07928e-7 8.91227e-8 3.67569e-8
1.91905e-10 2.59911e-8 6.30193e-8 1.47028e-7 3.30066e-7 7.12982e-7 1.48195e-6 2.96389e-6 5.70385e-6 1.05621e-5 1.88195e-5 3.22659e-5 5.32297e-5 8.44968e-5 0.000129063 0.000189689 0.000268261 0.000365047 0.000477986 0.000602225 0.000730093 0.000851675 0.000955973 0.00103251 0.00107304 0.00107304 0.00103251 0.000955973 0.000851675 0.000730093 0.000602225 0.000477986 0.000365047 0.000268261 0.000189689 0.000129063 8.44968e-5 5.32297e-5 3.22659e-5 1.88195e-5 1.05621e-5 5.70385e-6 2.96389e-6 1.48195e-6 7.12982e-7 3.30066e-7 1.47028e-7 6.30193e-8 2.59911e-8
2.12106e-10 1.76842e-8 4.2878e-8 1.00037e-7 2.24575e-7 4.85109e-7 1.00831e-6 2.01662e-6 3.88087e-6 7.1864e-6 1.28047e-5 2.19535e-5 3.62172e-5 5.74912e-5 8.78141e-5 0.000129063 0.000182523 0.000248376 0.00032522 0.000409751 0.000496752 0.000579475 0.000650439 0.000702513 0.000730093 0.000730093 0.000702513 0.000650439 0.000579475 0.000496752 0.000409751 0.00032522 0.000248376 0.000182523 0.000129063 8.78141e-5 5.74912e-5 3.62172e-5 2.19535e-5 1.28047e-5 7.1864e-6 3.88087e-6 2.01662e-6 1.00831e-6 4.85109e-7 2.24575e-7 1.00037e-7 4.2878e-8 1.76842e-8
2.32307e-10 1.15777e-8 2.80719e-8 6.54933e-8 1.47028e-7 3.17597e-7 6.60132e-7 1.32026e-6 2.54078e-6 4.70488e-6 8.38315e-6 1.43728e-5 2.37111e-5 3.76391e-5 5.74912e-5 8.44968e-5 0.000119497 0.00016261 0.000212919 0.000268261 0.00032522 0.000379378 0.000425837 0.00045993 0.000477986 0.000477986 0.00045993 0.000425837 0.000379378 0.00032522 0.000268261 0.000212919 0.00016261 0.000119497 8.44968e-5 5.74912e-5 3.76391e-5 2.37111e-5 1.43728e-5 8.38315e-6 4.70488e-6 2.54078e-6 1.32026e-6 6.60132e-7 3.17597e-7 1.47028e-7 6.54933e-8 2.80719e-8 1.15777e-8
2.52507e-10 7.29349e-9 1.76842e-8 4.12582e-8 9.26216e-8 2.00074e-7 4.15857e-7 8.31714e-7 1.60059e-6 2.96389e-6 5.28105e-6 9.0543e-6 1.49371e-5 2.37111e-5 3.62172e-5 5.32297e-5 7.52781e-5 0.000102438 0.00013413 0.000168994 0.000204875 0.000238993 0.000268261 0.000289738 0.000301113 0.000301113 0.000289738 0.000268261 0.000238993 0.000204875 0.000168994 0.00013413 0.000102438 7.52781e-5 5.32297e-5 3.62172e-5 2.37111e-5 1.49371e-5 9.0543e-6 5.28105e-6 2.96389e-6 1.60059e-6 8.31714e-7 4.15857e-7 2.00074e-7 9.26216e-8 4.12582e-8 1.76842e-8 7.29349e-9
2.72708e-10 4.42105e-9 1.07195e-8 2.50092e-8 5.61438e-8 1.21277e-7 2.52077e-7 5.04154e-7 9.70218e-7 1.7966e-6 3.20118e-6 5.48838e-6 9.0543e-6 1.43728e-5 2.19535e-5 3.22659e-5 4.56308e-5 6.2094e-5 8.13049e-5 0.000102438 0.000124188 0.000144869 0.00016261 0.000175628 0.000182523 0.000182523 0.000175628 0.00016261 0.000144869 0.000124188 0.000102438 8.13049e-5 6.2094e-5 4.56308e-5 3.22659e-5 2.19535e-5 1.43728e-5 9.0543e-6 5.48838e-6 3.20118e-6 1.7966e-6 9.70218e-7 5.04154e-7 2.52077e-7 1.21277e-7 5.61438e-8 2.50092e-8 1.07195e-8 4.42105e-9
2.92908e-10 2.57864e-9 6.2523e-9 1.4587e-8 3.27467e-8 7.07367e-8 1.47028e-7 2.94055e-7 5.65894e-7 1.04789e-6 1.86713e-6 3.20118e-6 5.28105e-6 8.38315e-6 1.28047e-5 1.88195e-5 2.66148e-5 3.62172e-5 4.74222e-5 5.97483e-5 7.24344e-5 8.44968e-5 9.48445e-5 0.000102438 0.000106459 0.000106459 0.000102438 9.48445e-5 8.44968e-5 7.24344e-5 5.97483e-5 4.74222e-5 3.62172e-5 2.66148e-5 1.88195e-5 1.28047e-5 8.38315e-6 5.28105e-6 3.20118e-6 1.86713e-6 1.04789e-6 5.65894e-7 2.94055e-7 1.47028e-7 7.07367e-8 3.27467e-8 1.4587e-8 6.2523e-9 2.57864e-9
3.13109e-10 1.44721e-9 3.50899e-9 8.18667e-9 1.83784e-8 3.96996e-8 8.25164e-8 1.65033e-7 3.17597e-7 5.8811e-7 1.04789e-6 1.7966e-6 2.96389e-6 4.70488e-6 7.1864e-6 1.05621e-5 1.49371e-5 2.03262e-5 2.66148e-5 3.35326e-5 4.06524e-5 4.74222e-5 5.32297e-5 5.74912e-5 5.97483e-5 5.97483e-5 5.74912e-5 5.32297e-5 4.74222e-5 4.06524e-5 3.35326e-5 2.66148e-5 2.03262e-5 1.49371e-5 1.05621e-5 7.1864e-6 4.70488e-6 2.96389e-6 1.7966e-6 1.04789e-6 5.8811e-7 3.17597e-7 1.65033e-7 8.25164e-8 3.96996e-8 1.83784e-8 8.18667e-9 3.50899e-9 1.44721e-9
3.33309e-10 7.81538e-10 1.89496e-9 4.42105e-9 9.92491e-9 2.1439e-8 4.45613e-8 8.91227e-8 1.71512e-7 3.17597e-7 5.65894e-7 9.70218e-7 1.60059e-6 2.54078e-6 3.88087e-6 5.70385e-6 8.06647e-6 1.09768e-5 1.43728e-5 1.81086e-5 2.19535e-5 2.56094e-5 2.87456e-5 3.1047e-5 3.22659e-5 3.22659e-5 3.1047e-5 2.87456e-5 2.56094e-5 2.19535e-5 1.81086e-5 1.43728e-5 1.09768e-5 8.06647e-6 5.70385e-6 3.88087e-6 2.54078e-6 1.60059e-6 9.70218e-7 5.65894e-7 3.17597e-7 1.71512e-7 8.91227e-8 4.45613e-8 2.1439e-8 9.92491e-9 4.42105e-9 1.89496e-9 7.81538e-10
3.5351e-10 4.0611e-10 9.84676e-10 2.29731e-9 5.15728e-9 1.11403e-8 2.31554e-8 4.63108e-8 8.91227e-8 1.65033e-7 2.94055e-7 5.04154e-7 8.31714e-7 1.32026e-6 2.01662e-6 2.96389e-6 4.19157e-6 5.70385e-6 7.46854e-6 9.40977e-6 1.14077e-5 1.33074e-5 1.49371e-5 1.61329e-5 1.67663e-5 1.67663e-5 1.61329e-5 1.49371e-5 1.33074e-5 1.14077e-5 9.40977e-6 7.46854e-6 5.70385e-6 4.19157e-6 2.96389e-6 2.01662e-6 1.32026e-6 8.31714e-7 5.04154e-7 2.94055e-7 1.65033e-7 8.91227e-8 4.63108e-8 2.31554e-8 1.11403e-8 5.15728e-9 2.29731e-9 9.84676e-10 4.0611e-10
3.73711e-10 2.03055e-10 4.92338e-10 1.14865e-9 2.57864e-9 5.57017e-9 1.15777e-8 2.31554e-8 4.45613e-8 8.25164e-8 1.47028e-7 2.52077e-7 4.15857e-7 6.60132e-7 1.00831e-6 1.48195e-6 2.09579e-6 2.85193e-6 3.73427e-6 4.70488e-6 5.70385e-6 6.65371e-6 7.46854e-6 8.06647e-6 8.38315e-6 8.38315e-6 8.06647e-6 7.46854e-6 6.65371e-6 5.70385e-6 4.70488e-6 3.73427e-6 2.85193e-6 2.09579e-6 1.48195e-6 1.00831e-6 6.60132e-7 4.15857e-7 2.52077e-7 1.47028e-7 8.25164e-8 4.45613e-8 2.31554e-8 1.15777e-8 5.57017e-9 2.57864e-9 1.14865e-9 4.92338e-10 2.03055e-10
3.93911e-10 9.76922e-11 2.3687e-10 5.52631e-10 1.24061e-9 2.67987e-9 5.57017e-9 1.11403e-8 2.1439e-8 3.96996e-8 7.07367e-8 1.21277e-7 2.00074e-7 3.17597e-7 4.85109e-7 7.12982e-7 1.00831e-6 1.3721e-6 1.7966e-6 2.26358e-6 2.74419e-6 3.20118e-6 3.5932e-6 3.88087e-6 4.03323e-6 4.03323e-6 3.88087e-6 3.5932e-6 3.20118e-6 2.74419e-6 2.26358e-6 1.7966e-6 1.3721e-6 1.00831e-6 7.12982e-7 4.85109e-7 3.17597e-7 2.00074e-7 1.21277e-7 7.07367e-8 3.96996e-8 2.1439e-8 1.11403e-8 5.57017e-9 2.67987e-9 1.24061e-9 5.52631e-10 2.3687e-10 9.76922e-11
4.14112e-10 0.0 1.09656e-10 2.55833e-10 5.74327e-10 1.24061e-9 2.57864e-9 5.15728e-9 9.92491e-9 1.83784e-8 3.27467e-8 5.61438e-8 9.26216e-8 1.47028e-7 2.24575e-7 3.30066e-7 4.66784e-7 6.35194e-7 8.31714e-7 1.04789e-6 1.27039e-6 1.48195e-6 1.66343e-6 1.7966e-6 1.86713e-6 1.86713e-6 1.7966e-6 1.66343e-6 1.48195e-6 1.27039e-6 1.04789e-6 8.31714e-7 6.35194e-7 4.66784e-7 3.30066e-7 2.24575e-7 1.47028e-7 9.26216e-8 5.61438e-8 3.27467e-8 1.83784e-8 9.92491e-9 5.15728e-9 2.57864e-9 1.24061e-9 5.74327e-10 2.55833e-10 1.09656e-10 0.0
4.34312e-10 0.0 0.0 1.13961e-10 2.55833e-10 5.52631e-10 1.14865e-9 2.29731e-9 4.42105e-9 8.18667e-9 1.4587e-8 2.50092e-8 4.12582e-8 6.54933e-8 1.00037e-7 1.47028e-7 2.07928e-7 2.82947e-7 3.70486e-7 4.66784e-7 5.65894e-7 6.60132e-7 7.40973e-7 8.00295e-7 8.31714e-7 8.31714e-7 8.00295e-7 7.40973e-7 6.60132e-7 5.65894e-7 4.66784e-7 3.70486e-7 2.82947e-7 2.07928e-7 1.47028e-7 1.00037e-7 6.54933e-8 4.12582e-8 2.50092e-8 1.4587e-8 8.18667e-9 4.42105e-9 2.29731e-9 1.14865e-9 5.52631e-10 2.55833e-10 1.13961e-10 0.0 0.0
4.54513e-10 0.0 0.0 0.0 1.09656e-10 2.3687e-10 4.92338e-10 9.84676e-10 1.89496e-9 3.50899e-9 6.2523e-9 1.07195e-8 1.76842e-8 2.80719e-8 4.2878e-8 6.30193e-8 8.91227e-8 1.21277e-7 1.58799e-7 2.00074e-7 2.42555e-7 2.82947e-7 3.17597e-7 3.43024e-7 3.56491e-7 3.56491e-7 3.43024e-7 3.17597e-7 2.82947e-7 2.42555e-7 2.00074e-7 1.58799e-7 1.21277e-7 8.91227e-8 6.30193e-8 4.2878e-8 2.80719e-8 1.76842e-8 1.07195e-8 6.2523e-9 3.50899e-9 1.89496e-9 9.84676e-10 4.92338e-10 2.3687e-10 1.09656e-10 0.0 0.0 0.0
4.74713e-10 0.0 0.0 0.0 0.0 9.76922e-11 2.03055e-10 4.0611e-10 7.81538e-10 1.44721e-9 2.57864e-9 4.42105e-9 7.29349e-9 1.15777e-8 1.76842e-8 2.59911e-8 3.67569e-8 5.00184e-8 6.54933e-8 8.25164e-8 1.00037e-7 1.16696e-7 1.30987e-7 1.41473e-7 1.47028e-7 1.47028e-7 1.41473e-7 1.30987e-7 1.16696e-7 1.00037e-7 8.25164e-8 6.54933e-8 5.00184e-8 3.67569e-8 2.59911e-8 1.76842e-8 1.15777e-8 7.29349e-9 4.42105e-9 2.57864e-9 1.44721e-9 7.81538e-10 4.0611e-10 2.03055e-10 9.76922e-11 0.0 0.0 0.0 0.0For the image metadata we specify the grid and the total flux of the image, which is 1.0. Note that we specify the total flux out front since it is degenerate with an overall shift in the gain amplitudes.
skymeta = (; mimg = mimg ./ flux(mimg), ftot = 0.6);We use again use a GMRF prior similar to the Imaging a Black Hole using only Closure Quantities tutorial for the log-ratio transformed image. We use the same correlated image prior for the inverse-logit transformed total polarization. The mean total polarization fraction p0 is centered at -2.0 with a standard deviation of 2.0 which logit transformed puts most of the prior mass < 0.8 fractional polarization. The standard deviation of the total polarization fraction pσ again uses a Half-normal process. The angular parameters of the polarizaton are given by a uniform prior on the sphere.
cprior = corr_image_prior(grid, dvis; order = 2)
skyprior = (
σs = ntuple(Returns(truncated(Normal(0.0, 0.5); lower = 0.0)), 4),
as = ntuple(Returns(cprior), 4),
)
skym = SkyModel(sky, skyprior, grid; metadata = skymeta)SkyModel
with map: sky
on grid:
RectiGrid(
executor: ComradeBase.Serial()
Dimensions:
(↓ X Sampled{Float64} LinRange{Float64}(-4.747133960864206e-10, 4.747133960864206e-10, 48) ForwardOrdered Regular Points,
→ Y Sampled{Float64} LinRange{Float64}(-4.747133960864206e-10, 4.747133960864206e-10, 48) ForwardOrdered Regular Points)
)
)Now we build the instrument model. Due to the complexity of VLBI the instrument model is critical to the success of imaging and getting reliable results. For this example we will use the standard instrument model used in polarized EHT analyses expressed in the RIME formalism. Our Jones decomposition will be given by GDR, where G are the complex gains, D are the d-terms, and R is what we call the ideal instrument response, which is how an ideal interferometer using the feed basis we observe relative to some reference basis.
Given the possible flexibility in different parameterizations of the individual Jones matrices each Jones matrix requires the user to specify a function that converts from parameters to specific parameterization f the jones matrices.
For the complex gain matrix, we used the JonesG jones matrix. The first argument is now a function that converts from the parameters to the complex gain matrix. In this case, we will use a amplitude and phase decomposition of the complex gain matrix. Note that since the gain matrix is a diagonal 2x2 matrix the function must return a 2-element tuple. The first element of the tuple is the gain for the first polarization feed (R) and the second is the gain for the second polarization feed (L).
function fgain(x)
gR = exp(x.lgR + 1im * x.gpR)
gL = gR * exp(x.lgrat + 1im * x.gprat)
return gR, gL
end
G = JonesG(fgain)Comrade.JonesG{typeof(Main.var"##225".fgain)}(Main.var"##225".fgain)Similarly we provide a JonesD function for the leakage terms. Since we assume that we are in the small leakage limit, we will use the decomposition 1 d1 d2 1 Therefore, there are 2 free parameters for the JonesD our parameterization function must return a 2-element tuple. For d-terms we will use a re-im parameterization.
function fdterms(x)
dR = complex(x.dRx, x.dRy)
dL = complex(x.dLx, x.dLy)
return dR, dL
end
D = JonesD(fdterms)Comrade.JonesD{typeof(Main.var"##225".fdterms)}(Main.var"##225".fdterms)Finally we define our response Jones matrix. This matrix is a basis transform matrix plus the feed rotation angle for each station. These are typically set by the telescope so there are no free parameters, so no parameterization is necessary.
R = JonesR(; add_fr = true)Comrade.JonesR{Nothing}(nothing, true)Finally, we build our total Jones matrix by using the JonesSandwich function. The first argument is a function that specifies how to combine each Jones matrix. In this case we will use the standard decomposition J = adjoint(R)_G_D*R, where we need to apply the adjoint of the feed rotaion matrix R because the data has feed rotation calibration.
js(g, d, r) = adjoint(r) * g * d * r
J = JonesSandwich(js, G, D, R)Comrade.JonesSandwich{Base.Splat{typeof(Main.var"##225".js)}, Tuple{Comrade.JonesG{typeof(Main.var"##225".fgain)}, Comrade.JonesD{typeof(Main.var"##225".fdterms)}, Comrade.JonesR{Nothing}}}(splat(js), (Comrade.JonesG{typeof(Main.var"##225".fgain)}(Main.var"##225".fgain), Comrade.JonesD{typeof(Main.var"##225".fdterms)}(Main.var"##225".fdterms), Comrade.JonesR{Nothing}(nothing, true)))Note
This is a general note that for arrays with non-zero leakage, feed rotation calibration does not remove the impact of feed rotations on the instrument model. That is, when modeling feed rotation must be taken into account. This is because the R and D matrices are not commutative. Therefore, to recover the correct instrumental terms we must include the feed rotation calibration in the instrument model. This is not ideal when doing polarized modeling, especially for interferometers using a mixture of linear and circular feeds. For linear feeds R does not commute with G or D and applying feed rotation calibration before solving for gains can mix gains and leakage with feed rotation calibration terms breaking many of the typical assumptions about the stabilty of different instrument effects.
For the instrument prior, we will use a simple IID prior for the complex gains and d-terms. The IIDSitePrior function specifies that each site has the same prior and each value is independent on some time segment. The current time segments are
ScanSeg()which specifies each scan has an independent valueTrackSeg()which says that the value is constant over the track.IntegSeg()which says that the value changes each integration time
For the released EHT data, the calibration procedure makes gains stable over each scan so we use ScanSeg for those quantities. The d-terms are typically stable over the track so we use TrackSeg for those.
intprior = (
lgR = ArrayPrior(IIDSitePrior(ScanSeg(), Normal(0.0, 0.2)); LM = IIDSitePrior(ScanSeg(), Normal(0.0, 1.0))),
lgrat = ArrayPrior(IIDSitePrior(ScanSeg(), Normal(0.0, 0.1))),
gpR = ArrayPrior(IIDSitePrior(ScanSeg(), DiagonalVonMises(0.0, inv(π^2))); refant = SEFDReference(0.0), phase = true),
gprat = ArrayPrior(IIDSitePrior(ScanSeg(), DiagonalVonMises(0.0, inv(0.1^2))); refant = SingleReference(:AA, 0.0), phase = true),
dRx = ArrayPrior(IIDSitePrior(TrackSeg(), Normal(0.0, 0.2))),
dRy = ArrayPrior(IIDSitePrior(TrackSeg(), Normal(0.0, 0.2))),
dLx = ArrayPrior(IIDSitePrior(TrackSeg(), Normal(0.0, 0.2))),
dLy = ArrayPrior(IIDSitePrior(TrackSeg(), Normal(0.0, 0.2))),
)(lgR = Comrade.ArrayPrior{Comrade.IIDSitePrior{Comrade.ScanSeg, Distributions.Normal{Float64}}, Base.Pairs{Symbol, Comrade.IIDSitePrior{Comrade.ScanSeg, Distributions.Normal{Float64}}, Tuple{Symbol}, @NamedTuple{LM::Comrade.IIDSitePrior{Comrade.ScanSeg, Distributions.Normal{Float64}}}}, Comrade.NoReference, Nothing}(Comrade.IIDSitePrior{Comrade.ScanSeg, Distributions.Normal{Float64}}(Comrade.ScanSeg(), Distributions.Normal{Float64}(μ=0.0, σ=0.2)), Base.Pairs{Symbol, Comrade.IIDSitePrior{Comrade.ScanSeg, Distributions.Normal{Float64}}, Tuple{Symbol}, @NamedTuple{LM::Comrade.IIDSitePrior{Comrade.ScanSeg, Distributions.Normal{Float64}}}}(:LM => Comrade.IIDSitePrior{Comrade.ScanSeg, Distributions.Normal{Float64}}(Comrade.ScanSeg(), Distributions.Normal{Float64}(μ=0.0, σ=1.0))), Comrade.NoReference(), false, nothing), lgrat = Comrade.ArrayPrior{Comrade.IIDSitePrior{Comrade.ScanSeg, Distributions.Normal{Float64}}, Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}, Comrade.NoReference, Nothing}(Comrade.IIDSitePrior{Comrade.ScanSeg, Distributions.Normal{Float64}}(Comrade.ScanSeg(), Distributions.Normal{Float64}(μ=0.0, σ=0.1)), Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}(), Comrade.NoReference(), false, nothing), gpR = Comrade.ArrayPrior{Comrade.IIDSitePrior{Comrade.ScanSeg, VLBIImagePriors.DiagonalVonMises{Float64, Float64, Float64}}, Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}, Comrade.SEFDReference{Float64}, Nothing}(Comrade.IIDSitePrior{Comrade.ScanSeg, VLBIImagePriors.DiagonalVonMises{Float64, Float64, Float64}}(Comrade.ScanSeg(), VLBIImagePriors.DiagonalVonMises{Float64, Float64, Float64}(μ=0.0, κ=0.10132118364233778, lnorm=-1.739120733481688)), Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}(), Comrade.SEFDReference{Float64}(0.0, 0), true, nothing), gprat = Comrade.ArrayPrior{Comrade.IIDSitePrior{Comrade.ScanSeg, VLBIImagePriors.DiagonalVonMises{Float64, Float64, Float64}}, Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}, Comrade.SingleReference{Float64}, Nothing}(Comrade.IIDSitePrior{Comrade.ScanSeg, VLBIImagePriors.DiagonalVonMises{Float64, Float64, Float64}}(Comrade.ScanSeg(), VLBIImagePriors.DiagonalVonMises{Float64, Float64, Float64}(μ=0.0, κ=99.99999999999999, lnorm=1.3823902436480708)), Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}(), Comrade.SingleReference{Float64}(:AA, 0.0), true, nothing), dRx = Comrade.ArrayPrior{Comrade.IIDSitePrior{Comrade.TrackSeg, Distributions.Normal{Float64}}, Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}, Comrade.NoReference, Nothing}(Comrade.IIDSitePrior{Comrade.TrackSeg, Distributions.Normal{Float64}}(Comrade.TrackSeg(), Distributions.Normal{Float64}(μ=0.0, σ=0.2)), Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}(), Comrade.NoReference(), false, nothing), dRy = Comrade.ArrayPrior{Comrade.IIDSitePrior{Comrade.TrackSeg, Distributions.Normal{Float64}}, Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}, Comrade.NoReference, Nothing}(Comrade.IIDSitePrior{Comrade.TrackSeg, Distributions.Normal{Float64}}(Comrade.TrackSeg(), Distributions.Normal{Float64}(μ=0.0, σ=0.2)), Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}(), Comrade.NoReference(), false, nothing), dLx = Comrade.ArrayPrior{Comrade.IIDSitePrior{Comrade.TrackSeg, Distributions.Normal{Float64}}, Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}, Comrade.NoReference, Nothing}(Comrade.IIDSitePrior{Comrade.TrackSeg, Distributions.Normal{Float64}}(Comrade.TrackSeg(), Distributions.Normal{Float64}(μ=0.0, σ=0.2)), Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}(), Comrade.NoReference(), false, nothing), dLy = Comrade.ArrayPrior{Comrade.IIDSitePrior{Comrade.TrackSeg, Distributions.Normal{Float64}}, Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}, Comrade.NoReference, Nothing}(Comrade.IIDSitePrior{Comrade.TrackSeg, Distributions.Normal{Float64}}(Comrade.TrackSeg(), Distributions.Normal{Float64}(μ=0.0, σ=0.2)), Base.Pairs{Symbol, Union{}, Tuple{}, @NamedTuple{}}(), Comrade.NoReference(), false, nothing))Finally, we can build our instrument model which takes a model for the Jones matrix J and priors for each term in the Jones matrix.
intmodel = InstrumentModel(J, intprior)InstrumentModel
with Jones: JonesSandwich
with reference basis: PolarizedTypes.CirBasis()intmodel = InstrumentModel(JonesR(;add_fr=true)) Putting it all together, we form our likelihood and posterior objects for optimization and sampling, and specifying to use Enzyme.Reverse with runtime activity for AD.
using Enzyme
post = VLBIPosterior(skym, intmodel, dvis)VLBIPosterior
ObservedSkyModel
with map: sky
on grid:
FourierDualDomain(
Algorithm: VLBISkyModels.NFFTAlg{Float64, AbstractNFFTs.PrecomputeFlags, UInt32}(1, 1.0e-9, AbstractNFFTs.TENSOR, 0x00000000)
Image Domain: RectiGrid(
executor: ComradeBase.Serial()
Dimensions:
(↓ X Sampled{Float64} LinRange{Float64}(-4.747133960864206e-10, 4.747133960864206e-10, 48) ForwardOrdered Regular Points,
→ Y Sampled{Float64} LinRange{Float64}(-4.747133960864206e-10, 4.747133960864206e-10, 48) ForwardOrdered Regular Points)
)
Visibility Domain: UnstructuredDomain(
executor: ComradeBase.Serial()
Dimensions:
189-element StructArray(::Vector{Float64}, ::Vector{Float64}, ::Vector{Float64}, ::Vector{Float64}) with eltype @NamedTuple{U::Float64, V::Float64, Ti::Float64, Fr::Float64}:
(U = -4.404687701333328e9, V = -4.521597269333328e9, Ti = 0.5833333432674408, Fr = 2.27070703125e11)
(U = -4.427688319999995e9, V = -4.558917845333328e9, Ti = 0.7333333566784859, Fr = 2.27070703125e11)
(U = -4.422283775999995e9, V = -4.545737898666661e9, Ti = 0.7333333566784859, Fr = 2.27070703125e11)
(U = -4.443824298666661e9, V = -4.596403783111106e9, Ti = 0.8833333253860474, Fr = 2.27070703125e11)
(U = -4.444612451555551e9, V = -4.594719701333328e9, Ti = 0.8833333253860474, Fr = 2.27070703125e11)
(U = -4.452459235555551e9, V = -4.700912867555551e9, Ti = 1.300000011920929, Fr = 2.27070703125e11)
(U = -4.453204522666661e9, V = -4.699246847999995e9, Ti = 1.300000011920929, Fr = 2.27070703125e11)
(U = -4.442525923555551e9, V = -4.738497991111106e9, Ti = 1.4500000029802322, Fr = 2.27070703125e11)
(U = -4.446617087999995e9, V = -4.727102591999995e9, Ti = 1.4500000029802322, Fr = 2.27070703125e11)
(U = -4.373388259555551e9, V = -4.848290346666661e9, Ti = 1.8916667103767395, Fr = 2.27070703125e11)
(U = -4.374057457777773e9, V = -4.846647935999995e9, Ti = 1.8916667103767395, Fr = 2.27070703125e11)
(U = 1.854972024888887e9, V = -4.900536604444439e9, Ti = 2.0416667461395264, Fr = 2.27070703125e11)
(U = -4.33647849244444e9, V = -4.885093333333328e9, Ti = 2.0416667461395264, Fr = 2.27070703125e11)
(U = 1.8543247111111093e9, V = -4.898899640888884e9, Ti = 2.0416667461395264, Fr = 2.27070703125e11)
(U = -4.337125802666661e9, V = -4.883456426666661e9, Ti = 2.0416667461395264, Fr = 2.27070703125e11)
(U = -6.191450481777771e9, V = 1.544323338368054e7, Ti = 2.0416667461395264, Fr = 2.27070703125e11)
(U = -4.1988920319999957e9, V = -4.985362616888884e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = 1.9771115377777758e9, V = -3.616703032888885e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = 2.112877287111109e9, V = -4.853924366222218e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = -4.1994733297777734e9, V = -4.983740188444439e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = 2.1122959964444423e9, V = -4.852301823999995e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = 1.9765302328888867e9, V = -3.6150805831111073e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = -6.311769287111104e9, V = -1.3143835066666654e8, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = -1.3576575533333316e8, V = 1.2372212337777765e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = -6.176003569777771e9, V = -1.368659580444443e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = 2.2020455187692285e9, V = -4.835182867692303e9, Ti = 2.612500011920929, Fr = 2.27070703125e11)
(U = 2.023443446153844e9, V = -3.5993280459487143e9, Ti = 2.612500011920929, Fr = 2.27070703125e11)
(U = 2.2014904845128183e9, V = -4.833565380923072e9, Ti = 2.612500011920929, Fr = 2.27070703125e11)
(U = 2.0228883987692285e9, V = -3.5977105394871755e9, Ti = 2.612500011920929, Fr = 2.27070703125e11)
(U = -1.7860208246153826e8, V = 1.2358548479999986e9, Ti = 2.612500011920929, Fr = 2.27070703125e11)
(U = 2.439166364444442e9, V = -4.776837418666661e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = 2.1385755875555534e9, V = -3.547005681777774e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = -3.9131562524444404e9, V = -5.122736583111106e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = 2.438692103111109e9, V = -4.775232839111106e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = -3.913630471111107e9, V = -5.121131932444439e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = 2.1381013582222202e9, V = -3.545401116444441e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = -6.352322559999993e9, V = -3.4589914133333296e8, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = -3.005907564444441e8, V = 1.229831726222221e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = -6.051731839999993e9, V = -1.575730865777776e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = 2.511586922666664e9, V = -4.755918350222218e9, Ti = 3.208333432674408, Fr = 2.27070703125e11)
(U = 2.170796600888887e9, V = -3.528796735999996e9, Ti = 3.208333432674408, Fr = 2.27070703125e11)
(U = 2.170351103999998e9, V = -3.5271960746666627e9, Ti = 3.208333432674408, Fr = 2.27070703125e11)
(U = 2.5111413973333306e9, V = -4.754317681777773e9, Ti = 3.208333432674408, Fr = 2.27070703125e11)
(U = -3.4079030488888854e8, V = 1.2271216071111097e9, Ti = 3.208333432674408, Fr = 2.27070703125e11)
(U = -3.554185649777774e9, V = -5.242140615111105e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = 2.691997553777775e9, V = -4.694789262222218e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = 2.2425047537777753e9, V = -3.4769514666666627e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = 2.2421424639999976e9, V = -3.4753602986666627e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = -3.5545479253333297e9, V = -5.24054941866666e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = 2.6916352782222195e9, V = -4.693198023111106e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = -4.4949279733333284e8, V = 1.2178377671111097e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = -6.246183196444438e9, V = -5.47351347555555e8, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = -5.796690360888883e9, V = -1.7651891164444425e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = 2.2622677727179465e9, V = -3.457387467487176e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = 2.7506836742564077e9, V = -4.671151025230764e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = -3.442546884923073e9, V = -5.272523263999994e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = -3.4428772102564063e9, V = -5.270935092512815e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = 2.2619374342564077e9, V = -3.455799302564099e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = 2.750353309538459e9, V = -4.66956284061538e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = -6.193230559179481e9, V = -6.013722551794866e8, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = -4.88415901538461e8, V = 1.2137635511794858e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = -5.704814683897429e9, V = -1.8151358063589723e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = 5.063708287999995e9, V = -4.2083771519999957e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 2.298526215111109e9, V = -3.400051128888885e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -3.088728042666663e9, V = -5.354635448888883e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 2.8947912462222195e9, V = -4.600179484444439e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 5.063473251555551e9, V = -4.206796074666662e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 2.894556095999997e9, V = -4.598598385777773e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 2.298291107555553e9, V = -3.398470051555552e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -3.0889631217777743e9, V = -5.35305430755555e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 2.1689170879999976e9, V = 3.918022879999996e8, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -5.983519260444438e9, V = -7.544559466666658e8, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -5.962650044444438e8, V = 1.2001283235555544e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -2.7651820871111083e9, V = 8.08326024888888e8, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -8.152436351999991e9, V = -1.1462582471111097e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -5.38725424355555e9, V = -1.954584263111109e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 5.20314781866666e9, V = -4.164995278222218e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 2.303663210666664e9, V = -3.3806049066666627e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -2.959920895999997e9, V = -5.380193535999994e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 2.934449208888886e9, V = -4.575548401777773e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 5.202945521777772e9, V = -4.163416078222218e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -2.96012315733333e9, V = -5.378614286222217e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 2.9342469404444413e9, V = -4.573969180444439e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 2.3034609279999976e9, V = -3.379025671111108e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 2.2686985813333306e9, V = 4.105531208888885e8, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -5.894370119111105e9, V = -8.046451235555547e8, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -6.307860088888882e8, V = 1.1949434986666653e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -2.8994846222222195e9, V = 7.843903822222214e8, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -8.163068671999991e9, V = -1.2151982364444432e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -5.26358409955555e9, V = -1.9995886115555534e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 5.54752803555555e9, V = -4.038701703111107e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 2.299188743111109e9, V = -3.3265329137777743e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 3.020574414222219e9, V = -4.505591765333328e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -2.5786923022222195e9, V = -5.445258367999994e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 5.547500060444439e9, V = -4.0388189439999957e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 5.545354601411759e9, V = -4.037995941647054e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 2.299206753882351e9, V = -3.325316984470585e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 3.0200180856470556e9, V = -4.504489261176466e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -2.5814593882352915e9, V = -5.443281573647054e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 5.545326531764701e9, V = -4.0381131670588193e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 2.5269535999999976e9, V = 4.668900488888884e8, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -7.213856906666659e8, V = 1.179058869333332e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -5.59926673066666e9, V = -9.396666328888879e8, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 2.5269256533333306e9, V = 4.667728097777773e8, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -3.248339306666663e9, V = 7.121688159999993e8, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -8.126220359111103e9, V = -1.406556682666665e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -4.877881031111106e9, V = -2.1187254968888867e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 3.248311303111108e9, V = -7.122860568888881e8, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 8.126192398222214e9, V = 1.4064394311111097e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 5.658309684512815e9, V = -3.9900336836923037e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 5.658283887589737e9, V = -3.990151154871791e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 3.0433165062564073e9, V = -4.479256681025637e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 2.290543504410254e9, V = -3.3066006843076887e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -2.4295591712820487e9, V = -5.467004468512815e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 5.658234788102558e9, V = -3.9884589423589706e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 3.0432416295384583e9, V = -4.477681939692303e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 2.2904686145641003e9, V = -3.305025916717945e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 5.658209004307686e9, V = -3.9885764069743543e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -2.429634061128203e9, V = -5.465429700923072e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 2.614993171692305e9, V = 4.8922299733333284e8, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -7.527729985641018e8, V = 1.1726559999999988e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -5.472875690666661e9, V = -9.87747757948717e8, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 2.6149673616410227e9, V = 4.891055195897431e8, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -3.367766186666663e9, V = 6.834330141538454e8, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -8.087868875487172e9, V = -1.4769707651282036e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -4.720102662564097e9, V = -2.160403784205126e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 3.3677403569230733e9, V = -6.835504902564095e8, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 8.087843052307683e9, V = 1.47685327753846e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 6.037485539555549e9, V = -3.7629412693333297e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 2.206725866666664e9, V = -3.2192776817777743e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 3.0838075448888855e9, V = -4.360287900444439e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 6.037469781333326e9, V = -3.763059541333329e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 6.037566023111104e9, V = -3.761366407111107e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 3.083888028444441e9, V = -4.358713059555551e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 2.206806371555553e9, V = -3.217702819555552e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 6.037550279111104e9, V = -3.7614846791111073e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 2.9536779662222195e9, V = 5.973466417777772e8, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = -8.77081679999999e8, V = 1.141010222222221e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 2.953662200888886e9, V = 5.97228362666666e8, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = -3.830759658666662e9, V = 5.436635751111104e8, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 3.8307439288888845e9, V = -5.43781858666666e8, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 6.193776597333326e9, V = -3.584673976888885e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 6.193768718222216e9, V = -3.584792625777774e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 2.0963110399999979e9, V = -3.1565621119999967e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 3.04824456533333e9, V = -4.2709149368888845e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 6.193972479999993e9, V = -3.583095103999996e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 2.0965068515555534e9, V = -3.154983210666663e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 3.0484403768888855e9, V = -4.269336042666662e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 6.193964600888882e9, V = -3.5832137315555515e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 3.145532039111108e9, V = 6.862409528888881e8, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 3.145524195555552e9, V = 6.861223146666659e8, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = -9.519335111111101e8, V = 1.114352821333332e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = -4.0974655644444404e9, V = 4.2811187466666627e8, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 4.0974577137777734e9, V = -4.2823050222222173e8, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 6.234329756444438e9, V = -3.385946503111108e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 1.931031367111109e9, V = -3.0921642239999967e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 6.234330837333326e9, V = -3.3860652231111073e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 2.9449131591111083e9, V = -4.1750833208888845e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 6.234647921777771e9, V = -3.3843593599999967e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 6.234648903111104e9, V = -3.3844781297777743e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 1.931349447111109e9, V = -3.090577116444441e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 2.94523125333333e9, V = -4.1734962275555515e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 3.289416654222219e9, V = 7.89136849777777e8, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 3.2894176639999967e9, V = 7.890181191111102e8, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = -1.0138817973333323e9, V = 1.0829191111111097e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = -4.303298439111107e9, V = 2.937822586666663e8, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 4.30329948444444e9, V = -2.939009928888886e8, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 6.151253688888882e9, V = -3.205430023111108e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 1.7430493191111093e9, V = -3.038615523555552e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 2.7939210951111083e9, V = -4.091441422222218e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 6.151262791111104e9, V = -3.205548579555552e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 6.159540360533327e9, V = -3.2139407530666633e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 2.804326297599997e9, V = -4.0944362495999956e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 6.159549064533327e9, V = -3.214059332266663e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 1.7550084863999982e9, V = -3.039883980799997e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 3.35733262933333e9, V = 8.86011391999999e8, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = -1.0508717404444433e9, V = 1.0528258951111101e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 3.357341724444441e9, V = 8.858928017777768e8, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = -4.408204373333328e9, V = 1.6681450088888872e8, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 4.408213447111106e9, V = -1.6693308977777758e8, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 6.086775295999993e9, V = -3.1345589279999967e9, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 2.719461119999997e9, V = -4.0595089759999957e9, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 1.6591904159999983e9, V = -3.0189068799999967e9, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 6.086787583999993e9, V = -3.1346773919999967e9, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 3.367314207999996e9, V = 9.24950019999999e8, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = -1.0602706839999988e9, V = 1.0406020399999988e9, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 3.367326447999996e9, V = 9.248315599999989e8, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = -4.427584863999995e9, V = 1.1565201249999988e8, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 4.427597119999995e9, V = -1.1577047599999988e8, Ti = 7.438888907432556, Fr = 2.27070703125e11)
)
)
ObservedInstrumentModel
with Jones: JonesSandwich
with reference basis: PolarizedTypes.CirBasis()Data Products: Comrade.EHTCoherencyDatumReconstructing the Image and Instrument Effects
To sample from this posterior, it is convenient to move from our constrained parameter space to an unconstrained one (i.e., the support of the transformed posterior is (-∞, ∞)). This transformation is done using the asflat function.
tpost = asflat(post)TransformedVLBIPosterior(
VLBIPosterior
ObservedSkyModel
with map: sky
on grid:
FourierDualDomain(
Algorithm: VLBISkyModels.NFFTAlg{Float64, AbstractNFFTs.PrecomputeFlags, UInt32}(1, 1.0e-9, AbstractNFFTs.TENSOR, 0x00000000)
Image Domain: RectiGrid(
executor: ComradeBase.Serial()
Dimensions:
(↓ X Sampled{Float64} LinRange{Float64}(-4.747133960864206e-10, 4.747133960864206e-10, 48) ForwardOrdered Regular Points,
→ Y Sampled{Float64} LinRange{Float64}(-4.747133960864206e-10, 4.747133960864206e-10, 48) ForwardOrdered Regular Points)
)
Visibility Domain: UnstructuredDomain(
executor: ComradeBase.Serial()
Dimensions:
189-element StructArray(::Vector{Float64}, ::Vector{Float64}, ::Vector{Float64}, ::Vector{Float64}) with eltype @NamedTuple{U::Float64, V::Float64, Ti::Float64, Fr::Float64}:
(U = -4.404687701333328e9, V = -4.521597269333328e9, Ti = 0.5833333432674408, Fr = 2.27070703125e11)
(U = -4.427688319999995e9, V = -4.558917845333328e9, Ti = 0.7333333566784859, Fr = 2.27070703125e11)
(U = -4.422283775999995e9, V = -4.545737898666661e9, Ti = 0.7333333566784859, Fr = 2.27070703125e11)
(U = -4.443824298666661e9, V = -4.596403783111106e9, Ti = 0.8833333253860474, Fr = 2.27070703125e11)
(U = -4.444612451555551e9, V = -4.594719701333328e9, Ti = 0.8833333253860474, Fr = 2.27070703125e11)
(U = -4.452459235555551e9, V = -4.700912867555551e9, Ti = 1.300000011920929, Fr = 2.27070703125e11)
(U = -4.453204522666661e9, V = -4.699246847999995e9, Ti = 1.300000011920929, Fr = 2.27070703125e11)
(U = -4.442525923555551e9, V = -4.738497991111106e9, Ti = 1.4500000029802322, Fr = 2.27070703125e11)
(U = -4.446617087999995e9, V = -4.727102591999995e9, Ti = 1.4500000029802322, Fr = 2.27070703125e11)
(U = -4.373388259555551e9, V = -4.848290346666661e9, Ti = 1.8916667103767395, Fr = 2.27070703125e11)
(U = -4.374057457777773e9, V = -4.846647935999995e9, Ti = 1.8916667103767395, Fr = 2.27070703125e11)
(U = 1.854972024888887e9, V = -4.900536604444439e9, Ti = 2.0416667461395264, Fr = 2.27070703125e11)
(U = -4.33647849244444e9, V = -4.885093333333328e9, Ti = 2.0416667461395264, Fr = 2.27070703125e11)
(U = 1.8543247111111093e9, V = -4.898899640888884e9, Ti = 2.0416667461395264, Fr = 2.27070703125e11)
(U = -4.337125802666661e9, V = -4.883456426666661e9, Ti = 2.0416667461395264, Fr = 2.27070703125e11)
(U = -6.191450481777771e9, V = 1.544323338368054e7, Ti = 2.0416667461395264, Fr = 2.27070703125e11)
(U = -4.1988920319999957e9, V = -4.985362616888884e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = 1.9771115377777758e9, V = -3.616703032888885e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = 2.112877287111109e9, V = -4.853924366222218e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = -4.1994733297777734e9, V = -4.983740188444439e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = 2.1122959964444423e9, V = -4.852301823999995e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = 1.9765302328888867e9, V = -3.6150805831111073e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = -6.311769287111104e9, V = -1.3143835066666654e8, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = -1.3576575533333316e8, V = 1.2372212337777765e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = -6.176003569777771e9, V = -1.368659580444443e9, Ti = 2.4583332538604736, Fr = 2.27070703125e11)
(U = 2.2020455187692285e9, V = -4.835182867692303e9, Ti = 2.612500011920929, Fr = 2.27070703125e11)
(U = 2.023443446153844e9, V = -3.5993280459487143e9, Ti = 2.612500011920929, Fr = 2.27070703125e11)
(U = 2.2014904845128183e9, V = -4.833565380923072e9, Ti = 2.612500011920929, Fr = 2.27070703125e11)
(U = 2.0228883987692285e9, V = -3.5977105394871755e9, Ti = 2.612500011920929, Fr = 2.27070703125e11)
(U = -1.7860208246153826e8, V = 1.2358548479999986e9, Ti = 2.612500011920929, Fr = 2.27070703125e11)
(U = 2.439166364444442e9, V = -4.776837418666661e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = 2.1385755875555534e9, V = -3.547005681777774e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = -3.9131562524444404e9, V = -5.122736583111106e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = 2.438692103111109e9, V = -4.775232839111106e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = -3.913630471111107e9, V = -5.121131932444439e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = 2.1381013582222202e9, V = -3.545401116444441e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = -6.352322559999993e9, V = -3.4589914133333296e8, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = -3.005907564444441e8, V = 1.229831726222221e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = -6.051731839999993e9, V = -1.575730865777776e9, Ti = 3.0583332180976868, Fr = 2.27070703125e11)
(U = 2.511586922666664e9, V = -4.755918350222218e9, Ti = 3.208333432674408, Fr = 2.27070703125e11)
(U = 2.170796600888887e9, V = -3.528796735999996e9, Ti = 3.208333432674408, Fr = 2.27070703125e11)
(U = 2.170351103999998e9, V = -3.5271960746666627e9, Ti = 3.208333432674408, Fr = 2.27070703125e11)
(U = 2.5111413973333306e9, V = -4.754317681777773e9, Ti = 3.208333432674408, Fr = 2.27070703125e11)
(U = -3.4079030488888854e8, V = 1.2271216071111097e9, Ti = 3.208333432674408, Fr = 2.27070703125e11)
(U = -3.554185649777774e9, V = -5.242140615111105e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = 2.691997553777775e9, V = -4.694789262222218e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = 2.2425047537777753e9, V = -3.4769514666666627e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = 2.2421424639999976e9, V = -3.4753602986666627e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = -3.5545479253333297e9, V = -5.24054941866666e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = 2.6916352782222195e9, V = -4.693198023111106e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = -4.4949279733333284e8, V = 1.2178377671111097e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = -6.246183196444438e9, V = -5.47351347555555e8, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = -5.796690360888883e9, V = -1.7651891164444425e9, Ti = 3.6249999403953552, Fr = 2.27070703125e11)
(U = 2.2622677727179465e9, V = -3.457387467487176e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = 2.7506836742564077e9, V = -4.671151025230764e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = -3.442546884923073e9, V = -5.272523263999994e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = -3.4428772102564063e9, V = -5.270935092512815e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = 2.2619374342564077e9, V = -3.455799302564099e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = 2.750353309538459e9, V = -4.66956284061538e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = -6.193230559179481e9, V = -6.013722551794866e8, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = -4.88415901538461e8, V = 1.2137635511794858e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = -5.704814683897429e9, V = -1.8151358063589723e9, Ti = 3.7791666984558105, Fr = 2.27070703125e11)
(U = 5.063708287999995e9, V = -4.2083771519999957e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 2.298526215111109e9, V = -3.400051128888885e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -3.088728042666663e9, V = -5.354635448888883e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 2.8947912462222195e9, V = -4.600179484444439e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 5.063473251555551e9, V = -4.206796074666662e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 2.894556095999997e9, V = -4.598598385777773e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 2.298291107555553e9, V = -3.398470051555552e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -3.0889631217777743e9, V = -5.35305430755555e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 2.1689170879999976e9, V = 3.918022879999996e8, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -5.983519260444438e9, V = -7.544559466666658e8, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -5.962650044444438e8, V = 1.2001283235555544e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -2.7651820871111083e9, V = 8.08326024888888e8, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -8.152436351999991e9, V = -1.1462582471111097e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = -5.38725424355555e9, V = -1.954584263111109e9, Ti = 4.225000083446503, Fr = 2.27070703125e11)
(U = 5.20314781866666e9, V = -4.164995278222218e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 2.303663210666664e9, V = -3.3806049066666627e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -2.959920895999997e9, V = -5.380193535999994e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 2.934449208888886e9, V = -4.575548401777773e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 5.202945521777772e9, V = -4.163416078222218e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -2.96012315733333e9, V = -5.378614286222217e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 2.9342469404444413e9, V = -4.573969180444439e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 2.3034609279999976e9, V = -3.379025671111108e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 2.2686985813333306e9, V = 4.105531208888885e8, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -5.894370119111105e9, V = -8.046451235555547e8, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -6.307860088888882e8, V = 1.1949434986666653e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -2.8994846222222195e9, V = 7.843903822222214e8, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -8.163068671999991e9, V = -1.2151982364444432e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = -5.26358409955555e9, V = -1.9995886115555534e9, Ti = 4.374999940395355, Fr = 2.27070703125e11)
(U = 5.54752803555555e9, V = -4.038701703111107e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 2.299188743111109e9, V = -3.3265329137777743e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 3.020574414222219e9, V = -4.505591765333328e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -2.5786923022222195e9, V = -5.445258367999994e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 5.547500060444439e9, V = -4.0388189439999957e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 5.545354601411759e9, V = -4.037995941647054e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 2.299206753882351e9, V = -3.325316984470585e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 3.0200180856470556e9, V = -4.504489261176466e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -2.5814593882352915e9, V = -5.443281573647054e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 5.545326531764701e9, V = -4.0381131670588193e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 2.5269535999999976e9, V = 4.668900488888884e8, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -7.213856906666659e8, V = 1.179058869333332e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -5.59926673066666e9, V = -9.396666328888879e8, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 2.5269256533333306e9, V = 4.667728097777773e8, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -3.248339306666663e9, V = 7.121688159999993e8, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -8.126220359111103e9, V = -1.406556682666665e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = -4.877881031111106e9, V = -2.1187254968888867e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 3.248311303111108e9, V = -7.122860568888881e8, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 8.126192398222214e9, V = 1.4064394311111097e9, Ti = 4.791666805744171, Fr = 2.27070703125e11)
(U = 5.658309684512815e9, V = -3.9900336836923037e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 5.658283887589737e9, V = -3.990151154871791e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 3.0433165062564073e9, V = -4.479256681025637e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 2.290543504410254e9, V = -3.3066006843076887e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -2.4295591712820487e9, V = -5.467004468512815e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 5.658234788102558e9, V = -3.9884589423589706e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 3.0432416295384583e9, V = -4.477681939692303e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 2.2904686145641003e9, V = -3.305025916717945e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 5.658209004307686e9, V = -3.9885764069743543e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -2.429634061128203e9, V = -5.465429700923072e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 2.614993171692305e9, V = 4.8922299733333284e8, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -7.527729985641018e8, V = 1.1726559999999988e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -5.472875690666661e9, V = -9.87747757948717e8, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 2.6149673616410227e9, V = 4.891055195897431e8, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -3.367766186666663e9, V = 6.834330141538454e8, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -8.087868875487172e9, V = -1.4769707651282036e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = -4.720102662564097e9, V = -2.160403784205126e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 3.3677403569230733e9, V = -6.835504902564095e8, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 8.087843052307683e9, V = 1.47685327753846e9, Ti = 4.945833384990692, Fr = 2.27070703125e11)
(U = 6.037485539555549e9, V = -3.7629412693333297e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 2.206725866666664e9, V = -3.2192776817777743e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 3.0838075448888855e9, V = -4.360287900444439e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 6.037469781333326e9, V = -3.763059541333329e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 6.037566023111104e9, V = -3.761366407111107e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 3.083888028444441e9, V = -4.358713059555551e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 2.206806371555553e9, V = -3.217702819555552e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 6.037550279111104e9, V = -3.7614846791111073e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 2.9536779662222195e9, V = 5.973466417777772e8, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = -8.77081679999999e8, V = 1.141010222222221e9, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 2.953662200888886e9, V = 5.97228362666666e8, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = -3.830759658666662e9, V = 5.436635751111104e8, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 3.8307439288888845e9, V = -5.43781858666666e8, Ti = 5.633333265781403, Fr = 2.27070703125e11)
(U = 6.193776597333326e9, V = -3.584673976888885e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 6.193768718222216e9, V = -3.584792625777774e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 2.0963110399999979e9, V = -3.1565621119999967e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 3.04824456533333e9, V = -4.2709149368888845e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 6.193972479999993e9, V = -3.583095103999996e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 2.0965068515555534e9, V = -3.154983210666663e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 3.0484403768888855e9, V = -4.269336042666662e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 6.193964600888882e9, V = -3.5832137315555515e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 3.145532039111108e9, V = 6.862409528888881e8, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 3.145524195555552e9, V = 6.861223146666659e8, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = -9.519335111111101e8, V = 1.114352821333332e9, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = -4.0974655644444404e9, V = 4.2811187466666627e8, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 4.0974577137777734e9, V = -4.2823050222222173e8, Ti = 6.1499998569488525, Fr = 2.27070703125e11)
(U = 6.234329756444438e9, V = -3.385946503111108e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 1.931031367111109e9, V = -3.0921642239999967e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 6.234330837333326e9, V = -3.3860652231111073e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 2.9449131591111083e9, V = -4.1750833208888845e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 6.234647921777771e9, V = -3.3843593599999967e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 6.234648903111104e9, V = -3.3844781297777743e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 1.931349447111109e9, V = -3.090577116444441e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 2.94523125333333e9, V = -4.1734962275555515e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 3.289416654222219e9, V = 7.89136849777777e8, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 3.2894176639999967e9, V = 7.890181191111102e8, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = -1.0138817973333323e9, V = 1.0829191111111097e9, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = -4.303298439111107e9, V = 2.937822586666663e8, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 4.30329948444444e9, V = -2.939009928888886e8, Ti = 6.716666579246521, Fr = 2.27070703125e11)
(U = 6.151253688888882e9, V = -3.205430023111108e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 1.7430493191111093e9, V = -3.038615523555552e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 2.7939210951111083e9, V = -4.091441422222218e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 6.151262791111104e9, V = -3.205548579555552e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 6.159540360533327e9, V = -3.2139407530666633e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 2.804326297599997e9, V = -4.0944362495999956e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 6.159549064533327e9, V = -3.214059332266663e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 1.7550084863999982e9, V = -3.039883980799997e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 3.35733262933333e9, V = 8.86011391999999e8, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = -1.0508717404444433e9, V = 1.0528258951111101e9, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 3.357341724444441e9, V = 8.858928017777768e8, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = -4.408204373333328e9, V = 1.6681450088888872e8, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 4.408213447111106e9, V = -1.6693308977777758e8, Ti = 7.233333349227905, Fr = 2.27070703125e11)
(U = 6.086775295999993e9, V = -3.1345589279999967e9, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 2.719461119999997e9, V = -4.0595089759999957e9, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 1.6591904159999983e9, V = -3.0189068799999967e9, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 6.086787583999993e9, V = -3.1346773919999967e9, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 3.367314207999996e9, V = 9.24950019999999e8, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = -1.0602706839999988e9, V = 1.0406020399999988e9, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 3.367326447999996e9, V = 9.248315599999989e8, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = -4.427584863999995e9, V = 1.1565201249999988e8, Ti = 7.438888907432556, Fr = 2.27070703125e11)
(U = 4.427597119999995e9, V = -1.1577047599999988e8, Ti = 7.438888907432556, Fr = 2.27070703125e11)
)
)
ObservedInstrumentModel
with Jones: JonesSandwich
with reference basis: PolarizedTypes.CirBasis()Data Products: Comrade.EHTCoherencyDatum
Transform: Params to ℝ^9788
)We can also query the dimension of our posterior or the number of parameters we will sample.
Warning
This can often be different from what you would expect. This difference is especially true when using angular variables, where we often artificially increase the dimension of the parameter space to make sampling easier.
Now we optimize. Unlike other imaging examples, we move straight to gradient optimizers due to the higher dimension of the space. In addition the only AD package that can currently work with the polarized Comrade posterior is Enzyme.
using Optimization, OptimizationLBFGSB
xopt, sol = comrade_opt(
post, LBFGSB();
initial_params = prior_sample(rng, post), maxiters = 2_000
)((sky = (σs = (3.3315032555918247, 0.19506460454000224, 0.07746759813226933, 0.016323814482378098), as = ((params = [0.006826322824203886 0.0135790871283861 0.020607387606374684 0.02907146328800041 0.040467809514302455 0.05445182202753945 0.0681472384405584 0.07937713120284347 0.08753742547711328 0.09237529321251635 0.09226630304496262 0.08453027414732561 0.07009454209024621 0.05266910694861659 0.03590662702423783 0.023138712359150514 0.015874095005949416 0.014858123583543173 0.01958540511948564 0.027366790812396576 0.03551096948200565 0.041992404802794196 0.04719496965373165 0.052441886851646004 0.05880076322876324 0.06659795460645722 0.07511769531459057 0.08163056500593249 0.08290134681409288 0.07770972339946022 0.06849196066047683 0.058670527642495134 0.049238578261609926 0.040280350348339246 0.033095852356068274 0.029214481275761466 0.030850373778213116 0.038820986781094954 0.05138368237169502 0.064766563576385 0.07450895415645796 0.07850351906322928 0.07703561754551576 0.07089402261613957 0.06042843130131666 0.04625150519305883 0.03049434507081566 0.015034605209912249; 0.015192269242944327 0.02954049900648445 0.04372740175030192 0.059955806978925254 0.0796966262602317 0.10213299894179156 0.12490570840138683 0.14567066585969832 0.16235634442339514 0.1713659820933557 0.1687197135285948 0.15472226277320947 0.13337652714226916 0.10888776361971336 0.08497807556084686 0.06446343026391729 0.050029548867745854 0.04448232962291197 0.04740024494798556 0.05593529095086305 0.06587813064072613 0.07423251951201798 0.08164858842210473 0.08988247705070415 0.10026644951188715 0.11277687475192896 0.1265461539890226 0.13791354250389792 0.1410552265741853 0.13235088376912715 0.11486709535000436 0.09588644121969764 0.08062198716018606 0.06977838805251996 0.06263791715414674 0.06024431636405474 0.064258432211574 0.07635357273988327 0.09601164763829334 0.12005719865706638 0.14255513279780901 0.15598977215052048 0.15545286030907582 0.14157174389937177 0.11857387200520642 0.09063171145524096 0.060567533070431626 0.030186605880999726; 0.023201720402507297 0.043599857499380966 0.06319515343177147 0.08611430200340009 0.11350559005106634 0.14406900948287094 0.17666308542497097 0.2091426757490671 0.23607817366017544 0.24960297676960227 0.24670598200638452 0.23202426704684959 0.21065265099722508 0.18577260170871665 0.15914988325134055 0.13265362670937303 0.11083731398375984 0.09691000889919801 0.0907006617612805 0.0904234834337594 0.09218868855087857 0.09450405040587471 0.09891669260462398 0.10779047212445934 0.12126119437035132 0.13777450193028565 0.15544297805292473 0.17125858483140752 0.1785327673310351 0.1711084019929174 0.1495638114334389 0.12280266272617361 0.10104522827765954 0.08856849056666131 0.0831928674114226 0.08396820999738464 0.09161757692005307 0.10706900093457569 0.13127469923724178 0.16262063007207705 0.19585313870975204 0.22084431997712248 0.22629877224225878 0.20876520379343746 0.17410013638820157 0.13128124893382176 0.08660685703935202 0.04278681439795374; 0.02786792390546406 0.05250208742173838 0.07725461201871102 0.10727733647460991 0.1438063878151573 0.1862450951348585 0.23391799985899042 0.2826057268328527 0.3225116565802086 0.34497270091874016 0.34906443837684203 0.33881511905752254 0.31926754358366105 0.2940597995054945 0.26478479293493906 0.23536929510704116 0.2096385496344737 0.18775339937857624 0.1695637285362254 0.15375429260045972 0.13978784611527076 0.1293681583072327 0.1263464119486255 0.1345711208559796 0.15235384236252814 0.17441522410809468 0.1949772090645591 0.21006389512401868 0.21472813289247816 0.20392945022223657 0.17743624238564845 0.1429347397398517 0.11255212454530006 0.09444038907398822 0.08865851912365044 0.09190395588979616 0.10502000294380971 0.12752632811863174 0.1579158775618926 0.19442720556847506 0.2317966571746086 0.26202047670880646 0.27420530107937935 0.25977193342495375 0.21985128122249872 0.16457247358211574 0.10602696876033431 0.05118150834558586; 0.0307491246660269 0.06103046174547021 0.09401420080228795 0.1327078817268878 0.17853552106558315 0.23375960724615522 0.2981445680899823 0.3641425479167691 0.41974841227300724 0.45569458833918675 0.46864452506460696 0.4622767671290292 0.44439212297666436 0.42081950429016757 0.396622699586865 0.37493468241193506 0.352359463537373 0.32510235410061467 0.29241737574226473 0.25639808276676557 0.2228664414378764 0.19663163127244224 0.18439259814950948 0.1909667111991921 0.21448050534850832 0.24501812605500214 0.2698808411613319 0.2790765357434749 0.26868580640673273 0.23896099215312167 0.19528041581711011 0.1464518227713246 0.10484620463389308 0.07998430397961635 0.0739903411144144 0.08187330518253524 0.10149662844009856 0.13343873773681217 0.1731889800385708 0.21583505282074317 0.25433378416811253 0.2811247731644126 0.2913319562172257 0.2787413712360179 0.239860963668548 0.18125483106293755 0.11655432427770578 0.05585779467416651; 0.035653496720154165 0.07560728366930991 0.12075187827869828 0.1693598463570144 0.22332763919086776 0.28841977857113366 0.36563260753449367 0.4463819291450455 0.5158009694686786 0.5597781623140076 0.5741171989654498 0.5672039026730207 0.5516842708670093 0.5398677456083333 0.539037450297731 0.5426705050590088 0.5355246582701243 0.504941867767528 0.4486501852881677 0.3808767749090405 0.3175380869255544 0.2687143105155018 0.24210222577386406 0.24206256835296577 0.2672581605837699 0.3059985791487504 0.339277779185191 0.3479931024123161 0.3236853806194411 0.27136058376641653 0.20411330023214236 0.135741982345173 0.07911598992410113 0.04627454297138646 0.040614579971259696 0.05620420144746023 0.08366011941375828 0.12204882970793848 0.16976557861739783 0.21936019338661586 0.2609815497189703 0.28297107316738973 0.2820150034239554 0.26193324694185893 0.22379835416232827 0.1704609317380503 0.11109029393966963 0.05386617084055534; 0.04579369214763096 0.09937473454738467 0.15772960380443452 0.2148955256447154 0.2746763246476311 0.34660902647896613 0.43319857761403796 0.5235782912622828 0.5969934295605294 0.6369895333376278 0.643415175943674 0.6301903646493152 0.6192186904875658 0.6315926161550559 0.6716159845467693 0.721895934186079 0.7487686511538277 0.7164977619200267 0.6216255176892206 0.4982031624147107 0.3824203902647681 0.2963866207570798 0.24678133214290046 0.23448255272632937 0.25487293334615146 0.29578541366305844 0.3361236527014318 0.35110799248281077 0.32541058512652266 0.2642840351196888 0.18540155522142773 0.10617428830406736 0.03877027112131653 -0.003205695221368245 -0.010554461614114326 0.013375604607900205 0.05336651961068701 0.09797423405336247 0.14720929961413548 0.19775003156786322 0.2401023626658239 0.26038078075082577 0.24999278686020626 0.21737758632103216 0.1753840853242464 0.12953424783475076 0.0833016125798188 0.040120450595223386; 0.06337146230597969 0.131131433429367 0.19786487461769806 0.2603370916510269 0.32753005325260737 0.41025836416992384 0.5074282790012116 0.6026981128973723 0.6725107214544032 0.7017996503320618 0.693125371504206 0.6676685929122369 0.6581043340555699 0.6902426585907101 0.769770623250572 0.8736069539670319 0.93972353915049 0.9022316967771872 0.7564814592804374 0.5558911035613939 0.36751786715523294 0.23269308934491387 0.15866779735650396 0.1374155358596945 0.15622922417707086 0.19899689882274163 0.24495204737563955 0.268517284210037 0.25160268403069846 0.1970249352889946 0.12422515308337877 0.049982008719103174 -0.015873074969367983 -0.06252358354581873 -0.07593881722433211 -0.04993738901343476 0.002906011910742175 0.05879294671863488 0.10766536194697414 0.15112308808778435 0.186437922454882 0.20342921331359254 0.19080079114166099 0.15224903489941244 0.1061997792251901 0.06629601280150592 0.03605963900803672 0.01503050878609881; 0.08380925512925763 0.163218695729739 0.23602156245551084 0.30944899119702357 0.3961167646005351 0.4998889484637884 0.6109097004259085 0.7093658590177983 0.7725126752451104 0.7864065642944691 0.7568394095945254 0.7119819640432417 0.6893142102416354 0.719111416545094 0.8136935262429523 0.943369960951732 1.0244623085569526 0.9746633824047735 0.7847063854003482 0.5162233401100484 0.2612324047521611 0.08233200263931591 -0.010193126469617744 -0.033634644891603886 -0.009469252115685584 0.04213794670861276 0.10021954116095622 0.1401458894056071 0.14142980455222331 0.10420073734324889 0.047713420575285936 -0.010900150612291047 -0.06707180408342527 -0.11260316860000887 -0.13465228153767403 -0.11941464338839618 -0.06627359046799094 -0.0010238478800956802 0.04960426204722421 0.08220716689214251 0.10404615568511129 0.11428342712956005 0.10354646381338493 0.06987903260242202 0.027665404882893194 -0.005189766968925995 -0.01955094093497533 -0.015478473445356473; 0.10040851664518387 0.19327610078727808 0.2833781321445059 0.38442715784844567 0.5032085797518796 0.6312379787280817 0.7521093080795215 0.8477057397685115 0.899014247065158 0.8944425329680793 0.841921106443105 0.7707986207219111 0.7205179088471547 0.7257129782631208 0.7996102370182772 0.9091232125228224 0.9717459824667075 0.9080475991132525 0.7007059393297431 0.40365404388242265 0.11601257934651106 -0.08490870031149303 -0.18540719585433563 -0.20686196385529973 -0.17676906732712108 -0.11314117571067904 -0.036137427190947743 0.03172394853152473 0.06394149144458429 0.05327487307965857 0.013980979596705688 -0.03164894657579129 -0.08050989571910594 -0.1289978928976431 -0.16253352968270188 -0.169124448761551 -0.13685406134617586 -0.07780796532143608 -0.02661767428679665 -0.0019458999858075908 0.004083917971216007 0.004196198611672372 -0.002804470110122973 -0.023567036158546995 -0.05231852380083605 -0.07276188397667721 -0.07104217935039696 -0.04384432845002684; 0.11714448472640021 0.23273781641298893 0.3554708305874627 0.49437267487680153 0.6436020414455225 0.7861919847432919 0.908274389024676 0.9985505857776484 1.041616477529215 1.0260619135141504 0.9555641813051541 0.8538153044804033 0.7622007492410157 0.7216121615995439 0.7487808679997673 0.8154717710756274 0.8463494342054225 0.7689320388226207 0.562468542132457 0.2694671273577923 -0.017234670689559035 -0.2163804450313613 -0.3087682582118575 -0.3205759822669476 -0.28095332892099867 -0.20573106673455194 -0.10884580262039127 -0.009389830157293404 0.06433175070886064 0.0909614060794537 0.07085712124152221 0.023816965737936447 -0.032962534124140075 -0.09833215326972812 -0.15415045146973289 -0.18733583055557967 -0.18967235184359788 -0.15824024014032376 -0.1176645855840257 -0.09659963034122777 -0.09899760330307454 -0.10865367436554528 -0.11361205470722287 -0.11819751370673391 -0.12663488666575215 -0.129345625027055 -0.11082980341081378 -0.06473437537107908; 0.14220303158874348 0.28958268330206416 0.45053971132277354 0.6242482448551314 0.7940305006729178 0.9433661430426381 1.0663290222767088 1.155234146619733 1.1974691162770985 1.1824135911919387 1.100042211215456 0.960465338867985 0.8109128362173749 0.7099980628356873 0.6883525857195898 0.7210366708378707 0.7290230541130825 0.6406422747161608 0.4389315674029156 0.1629172797181076 -0.107589013475744 -0.29114107734019173 -0.36343272541821386 -0.35572328639207806 -0.29766840512950704 -0.2133367104496243 -0.10527833205758266 0.014906396352863263 0.12643064152034555 0.19662527022591442 0.20590752807117266 0.15047090685921188 0.062393924922049294 -0.03621049758751503 -0.12746118658397634 -0.19153316839457007 -0.22609182848730652 -0.22894719412066705 -0.20994600028033053 -0.1944284566203237 -0.1965231993662297 -0.2074900997642176 -0.21036330632555233 -0.20159945916933517 -0.1885293112072007 -0.17055152679194677 -0.1358458180710796 -0.07665246380160458; 0.17569246582376255 0.3593570179733292 0.5566360190300867 0.7574876745157563 0.9397643863715552 1.0908242236827352 1.2080535925779465 1.2862532589130076 1.3227474592110198 1.3081025690167487 1.2169648168248015 1.0486160463935215 0.8509420169197802 0.6950817457247204 0.6295458742866707 0.6372714188122718 0.6378822515149275 0.5529315060377871 0.3643425125940691 0.1127961610307011 -0.1316720444670926 -0.29251451462223044 -0.342830984246302 -0.30766518370314566 -0.22697598824998147 -0.1388996430451289 -0.04035681591270363 0.074668540461133 0.1996432547567701 0.3046115637412186 0.348101680648872 0.2929770447137661 0.15956872872850306 0.011334079969325209 -0.1184710356864675 -0.21360129095294136 -0.27202620214126016 -0.29779779158765673 -0.2960163081162886 -0.2834962909630278 -0.27884054364870753 -0.28283878672710955 -0.28001688092792 -0.26132471733722396 -0.23176100875726294 -0.19530250541450767 -0.1465218784165457 -0.07990506521148899; 0.21276125716730546 0.4324407926421055 0.6604206029970773 0.8799009130970912 1.0661411611240381 1.2085941200945642 1.3058059234981196 1.35744511209218 1.370306757677595 1.3417163213369796 1.2519314932626409 1.0926391766283772 0.8868022867544297 0.6952792393638009 0.5845692659473709 0.5624819962599384 0.5627767291102768 0.4970076787845413 0.3339750326364506 0.11589480007822457 -0.09448717791178443 -0.2348849260858088 -0.26902984054479273 -0.20851884564688405 -0.10983308804325286 -0.019998673350172594 0.05447669472212901 0.13513313910564928 0.2340399262278431 0.34354246163788676 0.40907347019890766 0.36575930794961814 0.20410829388522864 0.003950187591798534 -0.16316929883768508 -0.2789189746419879 -0.34941152218261173 -0.3820517945993249 -0.382319675313763 -0.3624971472658879 -0.34151047736971546 -0.3297881099388384 -0.31784804288875185 -0.2919384270509739 -0.25183034273730764 -0.2030993419205428 -0.14559480582885534 -0.07692742881326652; 0.24694010170896727 0.49939088277574023 0.7527764649056058 0.9838425395393878 1.1666535745048177 1.292601632905932 1.3620090624446772 1.3775977670752164 1.3518304463214599 1.2951701955462782 1.2074564750265606 1.0798534075617876 0.9006867354358474 0.6999258329524476 0.55109606847871 0.4928729748213067 0.4856145305323624 0.4423453377867654 0.3187470717488993 0.1442268380545293 -0.03409103999313449 -0.1609581878776534 -0.1930055092664936 -0.13290426182892653 -0.032940982258138456 0.06083690096912379 0.12138260569265703 0.1567174910672268 0.1954441008309022 0.2571386349886559 0.3098375562902555 0.2779375715210591 0.13104378147721638 -0.07628966753335678 -0.26248209114100873 -0.38679383777217546 -0.4553190968155112 -0.4819624467321209 -0.47183248941797856 -0.43427711696143684 -0.3887308673508581 -0.35295847042576445 -0.3262233907196308 -0.2943862947232166 -0.2500495507309823 -0.1962610779167483 -0.136312293798974 -0.07041231457364489; 0.2768084895273114 0.5580192826832175 0.8327715028819704 1.071303899576225 1.2467555297366517 1.3533955488866545 1.3947621810489088 1.3748251801989957 1.3076844599902457 1.2143935276304496 1.1137433606874503 1.0050166463287578 0.8553174771106247 0.6625157283200945 0.49179309387795817 0.39905602790817374 0.37016351356601823 0.3407319749813461 0.26440235593200956 0.1399078586648424 -0.005487252949134179 -0.12107694125438123 -0.16634301423354192 -0.1332574230312099 -0.04731636609091323 0.05535804565255314 0.12403274056724421 0.11806283080711516 0.08264773108915205 0.05746727965076946 0.05183660505006461 0.017793124642761057 -0.09122160253140768 -0.25222019831131026 -0.4076534408882979 -0.5137849022297567 -0.5649519373734126 -0.57491116776631 -0.549715534529978 -0.493657850403682 -0.42315617570605 -0.36132753940438683 -0.31608322868578703 -0.27643359814552837 -0.2314583642459778 -0.17978670673335545 -0.12350482056567234 -0.06328695054006497; 0.30399031618087213 0.611221574122637 0.9056697475754085 1.1523248051408517 1.3224743685212659 1.4103550253420736 1.4202430729650553 1.358920334167383 1.2494357463523185 1.1279858706873718 1.0180160599640369 0.9084386594562921 0.7598730968740739 0.5676042939310625 0.38734832777661615 0.2699965721138765 0.21595474811332344 0.1933010387807384 0.1587220922438818 0.08268661374718679 -0.02584033337049584 -0.13195706838296728 -0.18853572949646036 -0.17551324057340842 -0.09090119291319564 0.03731434597485046 0.12224906968311137 0.07682067320331354 -0.04509030344614165 -0.16386862080424197 -0.2512746923593735 -0.31023360845490455 -0.38719456218152254 -0.49384961599342275 -0.593928097614642 -0.6520150067087175 -0.6628592039066412 -0.6414129655152295 -0.5953183358217916 -0.5242348295569669 -0.4375346256202452 -0.3571108887269877 -0.29635615576810126 -0.24899058561750415 -0.2042038136621211 -0.15814659832876996 -0.10980450689696955 -0.057071686071054624; 0.33150516639648886 0.6648373754913555 0.9808796911002442 1.2411040510630533 1.410506303734095 1.4747773730191167 1.436982418518967 1.3190538413166941 1.169265649595557 1.039179244539616 0.9400910593954547 0.8315060949392629 0.6655676689380805 0.4522721031425971 0.2538978897441361 0.11977783125742053 0.057979569251885 0.04898180807032325 0.048878797991344924 0.016186878338586543 -0.05955676599800484 -0.1503997539120378 -0.2011217646496688 -0.17656915962055708 -0.05113249344877766 0.12955400497424105 0.21385461459325228 0.12585877650029662 -0.09078998269050469 -0.310846615523487 -0.4765320601019498 -0.5798427374157802 -0.6504068579549576 -0.7161537352479521 -0.7696935347361789 -0.7847302609116094 -0.7507556814941927 -0.6861386695228173 -0.609424577109819 -0.5216595914904244 -0.42448457852276866 -0.3349570183685936 -0.2668428630092342 -0.2158545360948235 -0.1729884805382933 -0.13418535124784925 -0.09538490016443545 -0.050930845809075816; 0.3605232252985234 0.7218635065839243 1.0631733002749602 1.3411258172573348 1.5091519787096819 1.539954710433034 1.4395709598134943 1.2552203301644926 1.064702954275197 0.9309715083862464 0.8526314967456395 0.7622702197205385 0.5900908271017207 0.346304932604304 0.11189627633931663 -0.04205152379940777 -0.10285358969020941 -0.09975537410919742 -0.06550002349331034 -0.0398537398212834 -0.06470295772426829 -0.12776379051990888 -0.1657021875136212 -0.10264888903766856 0.08951503673637416 0.3260125580039974 0.40599887250032557 0.2789343444112088 0.0006250718289233306 -0.31501877713461396 -0.5653695873685305 -0.7233750397465075 -0.8107338623528343 -0.8564022855059811 -0.8764442867522947 -0.866943800700909 -0.8113365611620718 -0.716337729133435 -0.608579526496016 -0.5010262876053766 -0.3934480434734642 -0.2968755421747487 -0.22510339783160316 -0.1751659286290446 -0.1375152961080159 -0.10753956218314395 -0.07930577789356553 -0.04408074987925356; 0.3891863818422213 0.7786179394326762 1.1447183108989263 1.4372428230870709 1.595859438009824 1.5845659273545625 1.4231358050066591 1.1804465716622616 0.9507297833731626 0.8058461740496374 0.7451570611506622 0.6880709842626274 0.5332673549736934 0.26919885566195495 -0.007857531026057754 -0.19950085120693403 -0.28181215599426906 -0.2728040364249931 -0.18897406441275863 -0.08089834875902265 -0.021912419526409953 -0.037951428670172704 -0.058317520485952706 0.01748406315876834 0.21021352553697614 0.4358300206584845 0.536245530183694 0.42953456801245166 0.1441235082880671 -0.20003555299005896 -0.5047987131125444 -0.7230310174035948 -0.8404794948037003 -0.8778376552539793 -0.8794230711696941 -0.8658643113844052 -0.8160826170196637 -0.7196716614132705 -0.5997560866139515 -0.47845550467541864 -0.360616730536963 -0.25562092691345856 -0.17772248807657165 -0.12745512774604015 -0.09631859584574616 -0.07710873782533596 -0.0605446283909103 -0.03557852175495891; 0.41158644808591405 0.8231894788760499 1.2071703036240244 1.5034640988478443 1.6389738121651751 1.581575723734796 1.3743859331509916 1.09792115209863 0.8493291660758366 0.7049607502644478 0.6675559755887402 0.6481295494742145 0.5203874464081771 0.24867854021810407 -0.06761786367302361 -0.3085813981577385 -0.4271828844018718 -0.4217747910934439 -0.29950145397316214 -0.10043836222853642 0.07322701125148517 0.12933839503603103 0.10770371144492183 0.10989097528702191 0.18628891686475446 0.2810093170087474 0.34713785457300095 0.3331797016256399 0.15539302107838895 -0.11560393627912764 -0.3862951661033756 -0.6099265812479624 -0.7421791185895633 -0.7758071087790035 -0.7715201648371713 -0.772933935744424 -0.7532939208200714 -0.6830095862057346 -0.5759498359871249 -0.45773115041632295 -0.33752675091871565 -0.2250385580825078 -0.1375641890463191 -0.0819564939179513 -0.05336075438025988 -0.04325862235601528 -0.0386610193781604 -0.025282796903013875; 0.4209716438857698 0.8421110952001318 1.2321217438403789 1.5207515426769098 1.625813272922753 1.5274089854544932 1.2934319603732876 1.0121767991750148 0.7761320762476016 0.658356080908697 0.6561841322053021 0.6683217879409032 0.5538393217016774 0.2681101265351502 -0.0833080124629524 -0.36564314160991745 -0.5181718291042546 -0.5301112283922663 -0.38638092283181036 -0.10692751698638692 0.16848407460009737 0.31851481360933015 0.27218817719226646 0.12377623941866078 0.030477899218313176 0.007619927154153513 0.025852501973505467 0.053391341893634724 0.007438055671153129 -0.11738779217163922 -0.283182560875414 -0.4451245288874921 -0.5453133704937383 -0.5656690981253252 -0.5715146099770991 -0.6060830673930145 -0.6333862743950699 -0.6105304206697002 -0.5382748698867706 -0.43899216265694635 -0.3267540252138816 -0.2136103315807743 -0.11732333699561222 -0.051098543024677555 -0.018282993260063846 -0.0121316810808431 -0.016455853183220525 -0.013961799912264065; 0.41291044062942556 0.8270219314937272 1.2090356485576816 1.4844255535499522 1.569824803586575 1.4529461691270773 1.215555095729388 0.9531286590935721 0.7509834119733948 0.6681299294633705 0.6905167418650411 0.7128176218271495 0.590313982077725 0.2768227974259524 -0.11195764966990113 -0.4247768269469232 -0.5924692972352003 -0.6072304174755204 -0.44342304491398116 -0.11578251218975091 0.2321528142373828 0.45326907375059944 0.3343785921001235 0.03187717251211999 -0.19477238216775622 -0.2879234201386722 -0.26496046146139324 -0.17986594996164273 -0.11152583414435609 -0.1045888874429312 -0.15456361069780258 -0.23383244979762113 -0.28782944808024974 -0.2987912864644444 -0.3262250855899291 -0.39973090002284684 -0.4779291041076402 -0.5142563757770914 -0.49377354262231044 -0.4272426151587561 -0.33142827214417186 -0.22348822429066514 -0.12250137545676436 -0.04541253032804374 -0.002774060136382851 0.007057463395441495 -0.0001025686468282567 -0.004815989456940589; 0.387872946496821 0.7789141237184865 1.1404406376714848 1.4013419390949273 1.486367549947105 1.385195317848816 1.1774220444106696 0.9559306266549882 0.7908859557797996 0.7210429101705422 0.7287971945165346 0.7227222343416234 0.5713203675738556 0.22852769740330067 -0.195143701606967 -0.5329439939276132 -0.6953201987570176 -0.6834051950208587 -0.4937333778000045 -0.14117773454223867 0.2510678642681981 0.48560863579479524 0.276391769475692 -0.127850597562554 -0.4342173702238085 -0.5590270532572376 -0.5101797768768801 -0.3536829816746768 -0.18197476322489928 -0.06458063065929455 -0.016651865273653508 -0.024788993581823134 -0.043621624623803104 -0.06296824692188342 -0.11970565870291426 -0.21627904507073184 -0.323501639548168 -0.41075884950763014 -0.446946885666283 -0.42111639190789496 -0.34883080707202535 -0.25326202461000524 -0.1533391810019313 -0.06731864347079226 -0.012546115043521754 0.006952759313590075 0.004603160758360497 -0.0006145075641617087; 0.34866167589736385 0.7064247897662692 1.0431145462282185 1.2933057961840504 1.3946479353786447 1.3374873953441713 1.1859302273476144 1.0186208092444982 0.8839956261837437 0.7984634993112011 0.7453932165833757 0.6654865044966928 0.46990618848246485 0.11187819802434378 -0.33138649119709934 -0.6876059164895126 -0.8421925935116668 -0.8016229451512931 -0.5824710066848623 -0.2091062490916372 0.2046472343338126 0.4520228249831801 0.20614412899864834 -0.2447571851419035 -0.5945711898387553 -0.7368066794207392 -0.6860507735462241 -0.5039824652350626 -0.28334959157758693 -0.08725430907924744 0.04625797203424559 0.10297736415297007 0.10637937835076841 0.07160127691988642 -0.0007356635549997703 -0.0950345586690679 -0.20596168707625997 -0.32273722988168785 -0.40346884500614105 -0.41684565215373715 -0.37196356757502513 -0.2941278808508356 -0.20136886664316644 -0.11104396174705071 -0.04425471165235573 -0.01093541218243381 -0.002550930889295282 -0.0022538133436884773; 0.3013393546867594 0.6207052098731174 0.9349355647039437 1.1846882818094115 1.3163487488500956 1.3170230790184596 1.228380405134225 1.1107399997038663 0.995030578908628 0.8800091837630319 0.751110250025816 0.5855024487962066 0.34617129980678224 -0.006726585464925429 -0.4420081774667394 -0.8085302368216079 -0.9664940109264928 -0.9121400861933076 -0.6841130109650683 -0.30895595549340776 0.10952892086874666 0.38147087694417436 0.19308561985656653 -0.23821606090277309 -0.5948201247309192 -0.7505483580573633 -0.7238132279580861 -0.5690075140157359 -0.3580126738321705 -0.13618664215511642 0.04572234470573704 0.13924662640751653 0.14706823699628818 0.10468021519092989 0.04298914830011427 -0.027025663856272645 -0.12603341457314693 -0.2532602978660276 -0.3613992816899883 -0.4083897060678377 -0.39365540092120965 -0.33776643559273334 -0.2556117812744307 -0.1639878269087602 -0.08729697594994452 -0.04002323684933148 -0.01812821188288045 -0.008188581425561546; 0.2544916388614475 0.5347989648893455 0.8283408608341514 1.0842039502098806 1.251231241342869 1.3079799931623557 1.2748216407972361 1.1961085396321136 1.091428423346547 0.9505068929490229 0.7669911428471927 0.5430198060915046 0.27322261865084047 -0.06469699916141246 -0.46378126986410884 -0.8128689170994022 -0.9735979143447836 -0.9240382628621773 -0.7138954964017565 -0.37606178029261106 0.010536695282745078 0.286492500769614 0.21900495266638326 -0.12195463254819187 -0.4455123169024672 -0.6080014581997248 -0.6058491819671186 -0.48987182750268443 -0.3152884513127318 -0.11741479501844189 0.05723317681810703 0.1450658111361103 0.14514721319314436 0.10648234748298882 0.06599414587504152 0.01784284870912073 -0.07080596050593417 -0.19748656150811658 -0.31748507651336766 -0.39056084975787037 -0.4070452794233477 -0.3753016074576386 -0.30508791131607266 -0.21485804761747598 -0.13162559778523772 -0.07261704070295245 -0.03771496346865249 -0.01680236916575495; 0.21194754464352572 0.4562003009557809 0.7304623585367664 0.9912038072818697 1.1850227046728188 1.2813996581913345 1.2867851303324715 1.2341475893440983 1.1391141564180634 0.991256641995616 0.7916890773626448 0.5539086221756118 0.281953557891554 -0.036016671828573196 -0.3940079311624724 -0.7056776353678418 -0.8509903158708265 -0.8016526503595374 -0.6152721400426946 -0.3400899288754778 -0.02856298545717535 0.21594393448667634 0.2497545833252708 0.05027933438651017 -0.17923512271756709 -0.30741196365818685 -0.30936545607279614 -0.22740153794613732 -0.10954681325554096 0.022328615445959894 0.13968132326754462 0.18872704405746665 0.1711612286987778 0.13342122947940355 0.10198220560800371 0.05832534421372693 -0.02671160429069745 -0.14701137142426204 -0.27131517467826627 -0.3665367131458919 -0.41112937677696226 -0.3999392227819118 -0.3401244614843103 -0.25323879020766876 -0.16797477134991068 -0.10187646196538501 -0.05678753214812194 -0.0256844620501862; 0.17236495448754607 0.38304812298496344 0.6389565720934222 0.9014095876052401 1.1107085875299203 1.2253134209338592 1.243376075134407 1.1953311316159883 1.1035788402821285 0.966647687330203 0.7896867087677983 0.5847135095110177 0.3469309656706188 0.06412954541422489 -0.24949940472736926 -0.5198277240231965 -0.6441033521150882 -0.5863548791384571 -0.40550681106030023 -0.1818088562427835 0.03785087326876806 0.21102061181564877 0.28186433315059384 0.23230631492690418 0.15755823522772608 0.1274544471147512 0.14181798028134654 0.16861862088651178 0.18905656609955562 0.2103012973282704 0.22999900223307299 0.21964419846910482 0.18258849342070307 0.14852650257223607 0.12705740819215042 0.0913849220411546 0.015704639452975185 -0.09786251830922506 -0.22928205838690469 -0.3435119297270364 -0.4074975019996861 -0.40857358672233407 -0.3551942327238728 -0.2730301256761085 -0.19030100887034848 -0.12200650471694652 -0.07066975703703339 -0.03228273264625289; 0.13726506465885735 0.314860197963446 0.548342714155048 0.8065670410128466 1.0246439114447283 1.1464470725498923 1.1588243691999538 1.0921713713775565 0.9843592001827294 0.8537203686603623 0.7121944267137578 0.5671325088454019 0.4009308296521888 0.18450410600944925 -0.07519046936779109 -0.3079822891109692 -0.41889442007586214 -0.3642102098627091 -0.18820046186006362 0.006573575090122539 0.150021312043908 0.24066946922569205 0.2904947979092561 0.3192082274707296 0.3707128568557275 0.4468991449924934 0.4766253443466632 0.4300173427070765 0.3416816643047139 0.24913480766393853 0.175865632693111 0.12539052763871275 0.09882002301749235 0.10095601765196172 0.1182064333806192 0.11112636485926083 0.051550156262420876 -0.06147972941652767 -0.20395897258016654 -0.32965043556321405 -0.39876317899190666 -0.40024645424986693 -0.3490534064251715 -0.2732103874081579 -0.19674431534555367 -0.13069838409524287 -0.07746807897676694 -0.035504861153115054; 0.11201229677959013 0.25880586939375894 0.46219131809359654 0.704655858430763 0.9252007054286919 1.0547597391937036 1.0616871353281558 0.9699989870623926 0.8320117774796936 0.6885460656162392 0.5621638611732739 0.46059741147303895 0.3636958859946866 0.22989439266708206 0.04256895461828131 -0.1482187202682714 -0.2520975036926584 -0.21501436535635163 -0.06516907683081759 0.09960856536940745 0.19307987972907942 0.22208522704749065 0.23041346345105457 0.25313653966985517 0.3131553472768813 0.38340497008181545 0.39065365304428357 0.31074165349269345 0.18009368753447466 0.05021039890361422 -0.038921311473459105 -0.07591934771408397 -0.05953647245360175 0.003083830351778402 0.07529819775093405 0.1028744748571328 0.05440591733563669 -0.06306098006465796 -0.2083693922053309 -0.3263490927793552 -0.3824091282659149 -0.3749368874857172 -0.3240059333104831 -0.2558934025587664 -0.18883345583911046 -0.12907897429841023 -0.07789150435502183 -0.0357600172611272; 0.09751960195155153 0.22110918413618652 0.3924064797831678 0.6070102750958712 0.8191954316340933 0.9593751060327572 0.9771114396020106 0.8776940438971739 0.7148755425295157 0.5469452777406456 0.408785040294833 0.31100017058963225 0.23613411089635414 0.14522368436775787 0.01762354104784133 -0.1193680289098991 -0.20184409405466294 -0.177364885691588 -0.06085838572459018 0.06857661946972161 0.13504569746988335 0.13901943247776694 0.12049935949147807 0.10715493821722516 0.11278326269838822 0.11369896789138541 0.06997992930473257 -0.0321375271088319 -0.16355690343905996 -0.2693349053478416 -0.320229711008896 -0.3118372850685573 -0.24371341858243495 -0.13221513754473863 -0.022639286411971115 0.02755141945537143 -0.010354303108984067 -0.1167694921657257 -0.24020469824832033 -0.32861109884816014 -0.358080910003035 -0.33594985501072355 -0.2848292376833005 -0.22601989898624664 -0.17048545965159784 -0.11991595230098735 -0.07406958843858846 -0.03434923898694672; 0.09216569735269554 0.203489805411342 0.3478184834781397 0.5271915990324366 0.7179073063157481 0.8657399945114634 0.9123530166295607 0.8383890568312362 0.6791380503620259 0.4940532065123445 0.3289025789705272 0.20197938021392717 0.10297519571984129 0.002160071726137377 -0.1136833097480423 -0.2222345315626292 -0.28548404297233865 -0.26961358762814847 -0.17894966648214114 -0.07040889949768027 -0.007386029585349352 -0.0022228661160395833 -0.032432270488568324 -0.08159187816079962 -0.1378375883738552 -0.20658531923258527 -0.2976717804202231 -0.4163230890086957 -0.5335535645773836 -0.6050525619765562 -0.6159614163567627 -0.5676306117106868 -0.4681778183987365 -0.3335904600395215 -0.20335069325138527 -0.13184582576919512 -0.14257919007817518 -0.21116241756196824 -0.28710172725316246 -0.32794852899717064 -0.3232359747127759 -0.28750284970656115 -0.23932690538802853 -0.191013750580203 -0.1475302202464693 -0.10744092802444805 -0.06847321882309601 -0.03227281318254339; 0.0966665307536784 0.20571981861618255 0.3304226328384414 0.4730972345939503 0.6313379938590084 0.7769916256181567 0.8578437392733748 0.8343682864958385 0.7132262330543983 0.5376816625949532 0.3534833725731474 0.18848999770666808 0.046881998070029615 -0.08934785484175935 -0.23220867127599637 -0.3604197887139616 -0.4409524588275786 -0.45358593445563816 -0.398705557393492 -0.3112439025280642 -0.24734245002269079 -0.23793868040629207 -0.27760125092971055 -0.35240513958549485 -0.44711995544638944 -0.5511992020381505 -0.663473596985851 -0.784682313247973 -0.8886157758093842 -0.943822056704064 -0.9333438791721197 -0.8619411206895713 -0.742443424788957 -0.5892305136448011 -0.44100257876839893 -0.34327588185023816 -0.3099864896832678 -0.31750877172000924 -0.32684932125482746 -0.31356470268715 -0.279274259016801 -0.2368164019554374 -0.19490202867367723 -0.1576496682120508 -0.12576901836476248 -0.09534701587904333 -0.06293554347412557 -0.030379392761774905; 0.1120476757855325 0.22459534938827075 0.33492830059978795 0.44626655918754426 0.56990192811982 0.7025617283181583 0.808011552187418 0.838234974624109 0.7733388202054129 0.6334432040351297 0.45561254256849504 0.26989339365455534 0.09197489773696582 -0.08152437582751218 -0.2611638968115415 -0.43217475793960136 -0.5590929810559232 -0.617914268426751 -0.6075324513306049 -0.5516923424709932 -0.49948286506489514 -0.49620476974784117 -0.5502560241720795 -0.6442474745017019 -0.7590103374660975 -0.880439647319719 -1.005626414031433 -1.1279392011227978 -1.2255726580928932 -1.2685630997394075 -1.2394994957240182 -1.1490993520260424 -1.0107874156196939 -0.8435493809902606 -0.6820110013783593 -0.5552105567156438 -0.46837640528594304 -0.40554696958301156 -0.3475417025554495 -0.288095489071483 -0.23405559732114417 -0.191392828535068 -0.15830640257145098 -0.13170690645142488 -0.10900043922934466 -0.08559539792048063 -0.05838709961084983 -0.029027746677740617; 0.135334429628833 0.25433807875884046 0.3526404420372597 0.4396336912715433 0.535102272306112 0.6508362819238811 0.766382613775524 0.8367783832594275 0.8276126446907491 0.7368209327385788 0.587843673329616 0.40822127142488734 0.2163076772752556 0.01774942330445593 -0.18972160909690736 -0.39316621072850466 -0.5593961711467996 -0.6611817124228889 -0.6941517067803156 -0.6788224616290806 -0.655735731011502 -0.6708126661783927 -0.7431385669062712 -0.8591750318871335 -0.9962157507143865 -1.1388917359373196 -1.2791548301712528 -1.4034534897437037 -1.4907716393253356 -1.5150473945856553 -1.4708745267027283 -1.3704016633287652 -1.2259887225161619 -1.056322076957357 -0.8844037199390544 -0.7262789148638384 -0.5855222815723853 -0.45940273904821105 -0.34833396399274563 -0.25903856203231823 -0.19798228751428743 -0.16048103209730513 -0.13547256835703164 -0.11634845776264247 -0.09892786990718411 -0.07875492768926143 -0.05440833190084417 -0.027558242351438954; 0.157828259650853 0.2857845144293831 0.37691330344837126 0.4462770215043465 0.5196887686543797 0.6167816918060259 0.7290642987572211 0.820495372908458 0.8550946462067116 0.8170182990149711 0.7121580692206754 0.5604019656720266 0.3808708950112116 0.18208163367545893 -0.03264665197617288 -0.24777333893965559 -0.43061730945907684 -0.5561622922938257 -0.6220147997743734 -0.6480632324383873 -0.6680832643887695 -0.7173632893045544 -0.8162531717103902 -0.959037254517099 -1.1240619895119501 -1.2909019939068 -1.4433193398637767 -1.5658373848316525 -1.6399931244886399 -1.6522573349980554 -1.605304722666507 -1.5067396811337346 -1.3654293393725112 -1.1945975885352849 -1.0092139277506895 -0.82115495239364 -0.6390198940024019 -0.47350905098617707 -0.3371103837014663 -0.23995036286792137 -0.18149561461409597 -0.14849061776931632 -0.12772128238475755 -0.11204884745635214 -0.09551249956685243 -0.07474100630052431 -0.05091524370706795 -0.025773357531479864; 0.1689222370420436 0.3058089690837244 0.39971235620589696 0.46286384916398593 0.5206297471000637 0.5958056378076533 0.6906654165114641 0.7826772546713394 0.8417624763547709 0.8483664121913671 0.7956813340287301 0.6890135690049312 0.5437548728253131 0.3723109089266389 0.18057438248501173 -0.01792734854357228 -0.19516303821900108 -0.3279883112651625 -0.41589392010195586 -0.47558737736841034 -0.5328918228925054 -0.6130573816534959 -0.7333895911134377 -0.8938079240412423 -1.0780999495721635 -1.2633977464917119 -1.4280866873303333 -1.5558051194238016 -1.632516551558566 -1.6518254236949181 -1.6148978394510118 -1.5250656308215191 -1.3895421190717865 -1.219609895513743 -1.0281484168298036 -0.828223258456467 -0.634830688595516 -0.4642781095315603 -0.3299851600750507 -0.23891888350047175 -0.18529180215638164 -0.15440918807755138 -0.13436884569861088 -0.1174057607445315 -0.09769071624879548 -0.07421337440897115 -0.04948371329491625 -0.024773121374151556; 0.1637297966860292 0.303840089958593 0.40852576276909885 0.48076318106099747 0.5351977363073434 0.5900815964548205 0.6565879509071558 0.7304871850217721 0.7932590193399883 0.8265521445380193 0.8199517486916942 0.7673921924645091 0.6721844216218109 0.5474360841284718 0.40418377740858696 0.25285548477707204 0.10946361787232499 -0.011312817535592493 -0.10872924884952566 -0.19335789963220223 -0.2787227842598344 -0.37895764949761557 -0.5056295493422733 -0.6630292051694635 -0.8436513414347488 -1.0303857657286195 -1.203024050254165 -1.345047740517669 -1.4428863126098777 -1.487374146249584 -1.4743386283560511 -1.4057762575757122 -1.2896675261567212 -1.1368807151643734 -0.9603603421377662 -0.7757584464615849 -0.6005635804407737 -0.4495911713379039 -0.3333096317031727 -0.25531057268240737 -0.20769594754013374 -0.17703822887268658 -0.15303992628695395 -0.12979861605795284 -0.10454162983215724 -0.07771731707841045 -0.051068455313373805 -0.025263575325308953; 0.14675394802571592 0.28176831511177747 0.3964408552566762 0.48626353855671733 0.5513809592675373 0.5977972253250721 0.6378124136633273 0.6824822984431262 0.7304268662416566 0.7707369494433677 0.7938802841798289 0.7908817741378574 0.7538245638077945 0.6873636396854537 0.6049768295173878 0.5168558251335159 0.4293852523645549 0.3452715072016443 0.2618938371429591 0.17156236171401276 0.06811889426297826 -0.051700844459539194 -0.18888150568555 -0.34340690570168286 -0.5125098016266723 -0.6881241586980968 -0.8585502054234311 -1.0117377310764581 -1.133732824489975 -1.210304516249601 -1.232335949236791 -1.2000120830572187 -1.1193196821253095 -1.0001615713353043 -0.8563570463220412 -0.7054139698623898 -0.5641691138783437 -0.444309013165753 -0.3526901758566905 -0.28892833630377573 -0.2448519547696376 -0.21035591074661114 -0.1784809700138051 -0.14669479812226294 -0.11481344921038726 -0.08367349323062993 -0.05433242260821361 -0.02672126298708498; 0.1269959900947078 0.2516936826236659 0.3694178562819236 0.472863630836704 0.5533917594106503 0.6056919695798934 0.6351993109543561 0.6563074372091364 0.6804200509723188 0.7103232292262575 0.7437378843000393 0.7740552149964558 0.7899353672971795 0.7855346745251076 0.7680794212868193 0.7465167577043119 0.7225201917120861 0.6913945536948802 0.6442859954489036 0.5706170670505798 0.4635718400617264 0.3251650650684749 0.16469306280534318 -0.006259889091158033 -0.18019562910989495 -0.3528307086385029 -0.5216758706962686 -0.6824655785000069 -0.8239153818377447 -0.9298540367402877 -0.9886688154210138 -0.9964968120701498 -0.9557505085040691 -0.8756173543945157 -0.7703546714689858 -0.657258529368551 -0.5504544157498018 -0.45859634443135805 -0.38535004086711355 -0.3287926623630658 -0.2836127636998507 -0.24345425990483924 -0.20358824446132948 -0.16346586950593853 -0.12465614080171691 -0.08875816988276174 -0.056611723875179744 -0.027532305750313738; 0.11005828778047118 0.2235413133885241 0.3379214888842851 0.44488170984137876 0.5344351320288969 0.5981264015971434 0.6336150794348663 0.6484567440516855 0.656152278970134 0.6699707221199201 0.6986046593497748 0.7422957073063986 0.7925340923481757 0.8389846177792625 0.8792199752865106 0.9170497475382302 0.9515348708593762 0.9742397000403606 0.970899557598982 0.9251227107982047 0.8263068310154903 0.6773350653236259 0.4943446485193663 0.2984765684950462 0.10490482424157724 -0.08076071124792544 -0.25992729485738364 -0.4334205295728373 -0.5929269829965428 -0.723594296956862 -0.8132781276381588 -0.8549700221752533 -0.8484987811088894 -0.8014345420594631 -0.7272468810619198 -0.6412589815045202 -0.5551822968486242 -0.4773205073746093 -0.41160106951410197 -0.3572647127213613 -0.3108722544776219 -0.26651751462082696 -0.2200559675767314 -0.17297892847061022 -0.12873299975616714 -0.08933413990354355 -0.05544050459864047 -0.02634485704145585; 0.09605494947723146 0.1987124752452483 0.3058601420414648 0.40818143243187976 0.4975211493832204 0.5687669294569031 0.6166631245799837 0.6393150505153986 0.6444444677629884 0.6486574670486794 0.6685626713614182 0.712253407175895 0.7765886651613981 0.8515910753028473 0.9292543730602488 1.00705320099643 1.0816057475152372 1.1428938845131396 1.1742585500323703 1.155963876167707 1.073896901202293 0.9289570668106846 0.7388610360656738 0.5286240591785754 0.3176166474555647 0.11419854896148926 -0.08199475943601274 -0.2717726168457489 -0.4476513039831974 -0.5967484606102726 -0.7076298495495843 -0.7722161108047231 -0.7881135268156886 -0.7609036472338631 -0.7030328611726127 -0.6288860803503143 -0.5511157137124453 -0.479530780069923 -0.4182938264344263 -0.36658077716673987 -0.32024713622972645 -0.2727313190914576 -0.22144955921501816 -0.17059523094951148 -0.12433046794581129 -0.08402640602232941 -0.05043230913336198 -0.023254022372237133; 0.08268947479901453 0.17322995929522061 0.2699347067911083 0.36331736933027114 0.44635967050987485 0.5176922065289498 0.5744471557343573 0.6101004143799638 0.6237196079486625 0.6276720495970115 0.6412879749063296 0.6781707620189815 0.7406978685507604 0.8217179567996276 0.9119253019307592 1.0053925192680344 1.09634149899779 1.1742338630525437 1.2227153968805178 1.2229369697096901 1.1601697631462422 1.032438367231411 0.8542958798971246 0.6488837035235473 0.43547459364953406 0.2243872859656849 0.018644808226535137 -0.1798446522248474 -0.3629309772352495 -0.5188635361739323 -0.6375055911569846 -0.710923045845714 -0.7353558078970935 -0.7153255893711098 -0.6632423440947556 -0.5947047702657914 -0.5239115101979399 -0.45948990040353693 -0.40336145908877913 -0.35403550077625184 -0.3076493536236677 -0.25899589288893277 -0.20734434280112848 -0.15738803565177442 -0.11256532797128117 -0.07398606567188797 -0.04293651498763213 -0.019236535969072092; 0.06890547824041834 0.14437279127857042 0.22628543782081428 0.30770338245554874 0.3816536565475767 0.4466826574552232 0.503078411835259 0.5468303295759468 0.5727306876676502 0.5843257801373625 0.5962212749433413 0.6235601730117981 0.673127512212338 0.7424234579998206 0.8241380107218692 0.9120725432097851 0.999202354104725 1.0742315375337395 1.12240802032513 1.128155137274232 1.079440094598318 0.9733070913980683 0.8199594423466156 0.6369593686607024 0.4403528983092685 0.2408534585280302 0.044317907800083774 -0.144664570698002 -0.31718143053336845 -0.46267622504741024 -0.5727004414838648 -0.6403645183246328 -0.6619802070885392 -0.6419082535059405 -0.59271284423194 -0.530628802657011 -0.468769677412868 -0.4125877047780054 -0.3620921731277632 -0.3162321022373003 -0.27282415855281017 -0.2282539588837565 -0.1817461655899797 -0.13669430817690328 -0.09593677991932101 -0.061202730229069184 -0.03438225065539407 -0.015077300176749938; 0.055020865967412326 0.11353587419421352 0.17653314856607985 0.24146833853613384 0.30313136003524405 0.3579490064861609 0.40587083339665825 0.4464879051199548 0.47680353699838807 0.49574142540613114 0.5093100080845885 0.528901442676484 0.5629531584713492 0.6128297009794436 0.6749585097409412 0.7446127214696151 0.8151392064784232 0.8751841792315408 0.9116870525042439 0.9132724584260364 0.8722304001313901 0.7868032860652265 0.6636838888467693 0.5148807067666262 0.3521544436215002 0.18499522500556512 0.019960943598211482 -0.1372091862527136 -0.2785680303904941 -0.39625527186534587 -0.48407936158070186 -0.5364286443271009 -0.5501617050736167 -0.5293577095651176 -0.4852475440192259 -0.4321773582587816 -0.3808068480202154 -0.3347373856105621 -0.2934443723841878 -0.2560391630060289 -0.2210841176581728 -0.1853077866274091 -0.14740034095853077 -0.11007527915384382 -0.07603301460882106 -0.04724271696605151 -0.025740731465535426 -0.011069174717436235; 0.039949198044908554 0.08072042839840696 0.1230650418918244 0.16724894412868221 0.21140952317288275 0.25211124579425015 0.28729750453139163 0.31661014995507974 0.3397491083450248 0.35661112887010177 0.3690370585254175 0.38296962066096435 0.40501817595406 0.4373047787580802 0.47848169967980425 0.5259562762158336 0.574863100576254 0.6160350947239525 0.6388795338703915 0.6358227968037405 0.6027791812335325 0.5395016209386753 0.4506409743622566 0.3442781152204559 0.22833302189669644 0.10968933570316247 -0.006259063492721137 -0.11479963930691524 -0.21102505011455977 -0.2908163110179796 -0.3504432271077182 -0.38537894010406293 -0.3928367556241221 -0.37553138505869815 -0.34184343552699115 -0.30250154956114267 -0.26542308147771837 -0.23329760638253713 -0.20539584752040438 -0.18043151355716933 -0.156654914703524 -0.13143824530531617 -0.10422629240239265 -0.07741628954584373 -0.05294945943456485 -0.03230449007048615 -0.017209378018468845 -0.007292746500155012; 0.021521843384495934 0.0428448868809746 0.06422714034601883 0.08619468272249667 0.10882389090896276 0.13071800474752604 0.14980847935317657 0.16499451141664018 0.17631277606458484 0.18453634245187833 0.19116017415693687 0.19871862571758336 0.21038173918763645 0.22712216786391026 0.2481430048712062 0.2725475689073512 0.29787744535450267 0.3191621043289123 0.330341313336052 0.3273167729035495 0.30831699773496973 0.27364464339582417 0.22608775021829022 0.17001752754913427 0.10972640447101596 0.048756323585549266 -0.009925984538049359 -0.06390935642072458 -0.1112921586633173 -0.15090230874481977 -0.18112623037614087 -0.19905059415134632 -0.20272301396764222 -0.19328580619509558 -0.1752890340323359 -0.1545480184601683 -0.13545504004076822 -0.1194838102365833 -0.10591581646725286 -0.09369516768220912 -0.08151633373633704 -0.06813180065717561 -0.05373867509442691 -0.03975944940705488 -0.027051357118921746 -0.016313522722831934 -0.008571104180327787 -0.0036079757490100656], hyperparams = 95.60756438398377), (params = [-0.010615819984669157 -0.022214930563169193 -0.0348526526951253 -0.047812418063009565 -0.06023433811031251 -0.07157206551102488 -0.0808938355207471 -0.08523294020574133 -0.08088258020232772 -0.0658594167474067 -0.04069453294490219 -0.008092445801619426 0.02765206904034957 0.061881630932531254 0.09070396383780507 0.11134930376069437 0.12195664643762472 0.12244561383796815 0.1150711463107993 0.10340479732636412 0.0904448316002536 0.0771603797996852 0.06313563418106936 0.04725022381865988 0.02848665237306353 0.005187701621061604 -0.024174294455669616 -0.05919433616904175 -0.09634916617388124 -0.1308541140627241 -0.1590375362148253 -0.1791614420411132 -0.1913885882150099 -0.1963121800603594 -0.1945133715389886 -0.18772554960718854 -0.17674371786501406 -0.15960314569436823 -0.1339111280659618 -0.1005808549508258 -0.06380435899086306 -0.028014941920016006 0.0031597795124251584 0.026829316125256426 0.040742885993823016 0.043369079374086385 0.03527516162632222 0.019487394428487678; -0.019928273262239646 -0.04218260774815857 -0.067306169375141 -0.09373249725192828 -0.11866334365544942 -0.13941187137944042 -0.15300028239452718 -0.1547952629324762 -0.14020819158994152 -0.1076877769923924 -0.05948341588202762 -0.0006380208723445761 0.062034499610605136 0.12130852834411152 0.17088485533115097 0.20593115470077314 0.2234312637967 0.22341250138461066 0.20993965557758543 0.18943593460917388 0.1669649587108771 0.14421889581397118 0.11979471673078212 0.09081923915137521 0.05480741409755699 0.009252498123288756 -0.04773497440234186 -0.11455994531160853 -0.18463442528198346 -0.24926781979430254 -0.3019094288056093 -0.339656998780478 -0.36275573368530006 -0.3726234038516739 -0.3703819561877546 -0.3574977900597438 -0.33436484935876365 -0.2986436893386519 -0.24811957166393364 -0.18491330925823002 -0.11545502504033156 -0.04766110331988284 0.011008162617546214 0.054639213307454124 0.07962328127201031 0.08410760721315727 0.06889617387216296 0.0384544799107768; -0.026909476567115644 -0.057940827904019006 -0.09447710680183595 -0.13374850410789665 -0.16984872749377009 -0.19650605020745315 -0.20816322365249004 -0.20004726402492673 -0.1697950264415977 -0.11872563167586103 -0.05124672509524885 0.02581751184577428 0.1041340334609387 0.17543299860294864 0.2323855946587808 0.2697847113403655 0.28550310301634285 0.28121283002001596 0.26273206954088374 0.23748196183724815 0.2107309297911664 0.18393040761591262 0.15422867949594463 0.11678700926753678 0.06766925426488862 0.004533196534217025 -0.07348555953804775 -0.16313525839721785 -0.2558359955665878 -0.34101306958380023 -0.41086582190495813 -0.4621956179760632 -0.49522969553648455 -0.5113896572149025 -0.5108659396664506 -0.49275960090016746 -0.4560224515653616 -0.3999362504566184 -0.32558985162533105 -0.23722397759496228 -0.14200955209557098 -0.049377767289123346 0.03055724496458833 0.08894956241722607 0.12066582734238528 0.12428448618359063 0.10133317606976898 0.05678252763158428; -0.030760935235294332 -0.06755375252592931 -0.11244677020028482 -0.16111083838722223 -0.20431229363994077 -0.23218033414264955 -0.23728915272094322 -0.21630023387965938 -0.17053332599735907 -0.10432244406121687 -0.024124561770919983 0.06126652659504402 0.1423455480603322 0.21049458598756549 0.2590290360109866 0.285391565191191 0.29122122874834594 0.2815638014812417 0.26323505449970885 0.24141535402334535 0.21847861258324944 0.19347113474925132 0.16226279121550935 0.11953161923974542 0.06140894560237683 -0.013296285436871625 -0.103604766589945 -0.20499691614096593 -0.3083239373395258 -0.4029316375532499 -0.48104323188805653 -0.5396631830167313 -0.5787727407073368 -0.5991223709365321 -0.5995172124438165 -0.5763992743199277 -0.526832043992768 -0.45127466147999484 -0.35476356680344434 -0.24507749010444346 -0.1309934079730372 -0.02231221311349742 0.07059913400682716 0.13747320837343271 0.1710018262093876 0.16932803949002492 0.13530529781491588 0.07520749896682483; -0.03128305209800454 -0.06972204006085542 -0.11764395906746243 -0.16947179855765612 -0.21352015292854212 -0.23763950409365509 -0.2341000798992422 -0.20193407952092532 -0.14575264335884391 -0.0727218333029789 0.008171832384015289 0.08694560909602474 0.15444520157658762 0.2037394165203123 0.23224770311234164 0.24264466976360505 0.2411546741140264 0.23533230192439358 0.2297932896595433 0.22409992163361125 0.21479366984583406 0.196642723159097 0.16468394383558796 0.11556089554312159 0.04855143025496408 -0.03476975971144726 -0.1316819254761268 -0.23700531216404744 -0.3425069891187556 -0.43837243521630054 -0.5172971886439777 -0.5759751666175701 -0.6138883789915179 -0.6309054029216097 -0.6261161580660813 -0.5956015171154554 -0.5355317210097299 -0.44551431667650243 -0.33139838059510796 -0.20432420324700762 -0.07631643144061782 0.04107740789545217 0.13784864968781543 0.20489698935789027 0.23408109929247786 0.22220653467084373 0.17299393436574248 0.09476316250123054; -0.02870083031876619 -0.06436646580909122 -0.10921032447209746 -0.15717560586159543 -0.19572097503823466 -0.21240177625790968 -0.20045520908474462 -0.16130944367872577 -0.1027748493467515 -0.035233448580140894 0.03126357825887447 0.08826064099184155 0.1297132826438391 0.15426278976830343 0.16607130631578898 0.17197861536986542 0.17962879804633355 0.19415289079503348 0.21384408814047784 0.2320254924263986 0.23937889007033647 0.22703404565867225 0.19000117874099964 0.12830758664367387 0.0468526145728898 -0.04795742002885432 -0.1507932324913725 -0.25621982782406993 -0.3581326443618209 -0.4488476602386159 -0.5220293781627929 -0.5740626930822262 -0.6039449011732648 -0.6104615840727663 -0.5931406634592811 -0.5505335336526488 -0.4801432986918088 -0.3813811672888744 -0.25767816444747377 -0.1199756398753009 0.016932930460685985 0.13817444802391457 0.23186651622070165 0.2905902722824736 0.3084186737972767 0.28181711061327663 0.21424594107169062 0.11563298538208702; -0.023171435089892705 -0.0523970060512897 -0.08923802282783815 -0.12759792074458393 -0.15587048721491725 -0.16298496034388385 -0.14404295277887494 -0.1033310062241979 -0.05149300020823825 0.00039669098770521135 0.04443580798628265 0.07600816547751003 0.09528309404478758 0.10856364158113216 0.12441922569605202 0.1491875406290255 0.1858866692647495 0.23181474526005338 0.27861339972445415 0.31509314221679935 0.32906082415170435 0.3116487736531793 0.2587749701790219 0.17412340206417284 0.06828945516879029 -0.046618138804540916 -0.16027650866945978 -0.2665526367709933 -0.36144934024273184 -0.4415792473501498 -0.5025934618111806 -0.5416164277698079 -0.5573998572380577 -0.5488014608456858 -0.5143521371324445 -0.45484723884000827 -0.37099632555121975 -0.26493453405703193 -0.1391177325805825 -0.00027705360648431056 0.13759314690891158 0.2574460583092191 0.344187014737178 0.38954333822957443 0.390557605797574 0.34569678753335886 0.25795426523433357 0.1376390234640961; -0.015391494298439294 -0.03548441646922484 -0.060947985148101645 -0.08611383976439727 -0.10105294209694585 -0.09706539171392958 -0.07254703242354066 -0.03430556530936756 0.0071941181191214915 0.04358387662448893 0.07099453583522829 0.09048325132799766 0.10920321648424357 0.1376412351015742 0.18269231469752406 0.24401871811162498 0.31525530165291726 0.38786245247148327 0.4513235255504479 0.49071384290699477 0.4940831925805904 0.4554438953359498 0.3734577635928887 0.255631634254216 0.11597124073004061 -0.028154936331584675 -0.16078705238874522 -0.2728564310192108 -0.3615460551464523 -0.42825308158999975 -0.47290803075506427 -0.4942899900884139 -0.49132088991712497 -0.4637164816946536 -0.4100061872634485 -0.3310091243712074 -0.2301012086710807 -0.11303205143812263 0.014021849251428528 0.1466046619315825 0.2758404661750859 0.386829317915015 0.46307534847148146 0.49359723583578907 0.47586031974928467 0.4110060485511572 0.30259760819560916 0.16038682940874002; -0.006185914089346551 -0.015312358036617601 -0.027274075412629786 -0.036999582512912466 -0.036130938713530354 -0.01918757556157252 0.011508073005980625 0.04838189105543643 0.08385429955245453 0.1140787218294903 0.14045581809626073 0.1700068904262795 0.21404130030933327 0.28138371971397746 0.3701340317310327 0.4676495119281709 0.5606281344372325 0.6431800237852745 0.7056890791192337 0.731098704940494 0.7086188769634061 0.6349379352119561 0.5147959789235278 0.35911018936006106 0.18256893214328676 0.005361194591510642 -0.1533273647422707 -0.2789514673916566 -0.3677891030314698 -0.42255873919723436 -0.44898703015514113 -0.44953510530069746 -0.42468052290308034 -0.37486929247725687 -0.30034463002611156 -0.20213740117239806 -0.08397589001867407 0.047098995540529105 0.1804935472825606 0.3077993556758355 0.4232619910164914 0.5184181538266476 0.579849435709111 0.5948381083651607 0.558930616836697 0.474398433632556 0.34588576885973094 0.18258097579355662; 0.004895285516143444 0.008648312744430252 0.012003852892917305 0.01977656460092177 0.0386976446406449 0.07029938091820527 0.10906795688981692 0.14841882486164515 0.18481571269821404 0.21910987086844058 0.2575179526379318 0.3121355129350171 0.3962663367238997 0.5126410124545854 0.6458596323104391 0.7710247438720528 0.8727214844833608 0.9483758875837616 0.9913052501047599 0.988219344729181 0.9289638072997827 0.8140252356927838 0.6556943394228527 0.4667888354832606 0.26127313667244606 0.05642524689767216 -0.12928163680936272 -0.2776609576902814 -0.37964345391226617 -0.4341604746920314 -0.4471533409228029 -0.42657513706751826 -0.37682706365472635 -0.30167235103237666 -0.20388993942705802 -0.08629304410633046 0.0473384283364685 0.19123058786841013 0.33430820106056475 0.4631611585079445 0.5686202811866689 0.6465051579474135 0.6903414934631987 0.6889518012885645 0.6358074214923447 0.5329129862083093 0.3856743788078016 0.20292032660027134; 0.019190992515667894 0.03851284126036969 0.05921456750008271 0.08594223843647851 0.12336089944168586 0.16961686697362682 0.2182708428351259 0.26452145975069125 0.3064969605944549 0.3471005163296723 0.3970536902623138 0.4742695307470415 0.5935197371918868 0.7512316242898924 0.9216064637111359 1.0713729215050123 1.1782109473606404 1.2358236862710423 1.2452841967086863 1.2022551094179608 1.100056305051202 0.9472978163377704 0.7620623046357659 0.5573920757297844 0.343662297494012 0.12936990269327742 -0.07271904369927544 -0.24691350028864456 -0.37606903939590924 -0.45146052425795374 -0.46943431431100013 -0.4373062198037745 -0.3644164834659693 -0.26132819685354886 -0.13757410880300316 0.0016819972054767766 0.15166309527033295 0.3073948889420061 0.4600723560912467 0.5953992025535685 0.6991927604106497 0.7651550854023674 0.792166554473253 0.774364701099091 0.7049559139044712 0.5849365871073567 0.42048939411223846 0.22053649566936737; 0.03699180824953424 0.07449487314850509 0.11404480534000458 0.15962049935854103 0.2141591567792765 0.2754623609818812 0.33815916550923997 0.3955386682710607 0.44181808834187225 0.48015695034844336 0.5267261402400154 0.606265798912187 0.736429811138026 0.912595244757205 1.1062434559043697 1.276858227815684 1.3883389375923474 1.4268057590932288 1.3992729513632867 1.3106931224211984 1.166444882336099 0.9883015464305033 0.7986648820005203 0.6068896705525122 0.41426275331780354 0.21694538218749654 0.01709745903666078 -0.1750541959224624 -0.3379523337002049 -0.45175382772896866 -0.499815905587329 -0.4767919951615472 -0.3927168588139096 -0.26410642058900163 -0.1122966913252568 0.05062949101244551 0.22005959823161025 0.3904355860007047 0.5536473940509001 0.6974620306444772 0.8057469981888774 0.8674669569758593 0.8820956798643886 0.8494062461456541 0.765232336706661 0.629926361633608 0.44996448502455527 0.23504827948416898; 0.05738761513763937 0.1148380345472791 0.17361728232733142 0.23705897471082626 0.308403479786398 0.3876487972993103 0.469098264008115 0.5392096259124816 0.5845421741763474 0.6060921507006958 0.6253139538603372 0.6766342395824964 0.7846926551610529 0.9489247024359907 1.1430873253426577 1.3222156461066654 1.439157453388043 1.468787180636276 1.4121867167782531 1.2832058090952574 1.1101063563326334 0.9319916630681526 0.7695246613143788 0.6216536851291155 0.4765862222145876 0.3168705739890662 0.13337092871417405 -0.0662668099233081 -0.26121072437752935 -0.4210387219248772 -0.5168359978094917 -0.5260416510215176 -0.45178492983820134 -0.311934412469125 -0.1346314273158346 0.054156575386933865 0.2455551739734238 0.43508690927960686 0.6128470901032134 0.7671545311408945 0.8831625997237201 0.9466243256300001 0.9548583858183437 0.9111076589023044 0.8148415629365715 0.6668973401909789 0.47397591958658813 0.2466860119798231; 0.07928544266784898 0.1571208186230117 0.23419808659390914 0.31415703909116116 0.40199642445755346 0.500448414474183 0.6029279419656782 0.6878658934163082 0.7308569679007291 0.7259046695077691 0.696541695221875 0.6890392482502093 0.7430507425375489 0.8663145240253289 1.0350632728549902 1.2058520409094304 1.3282603250345077 1.3622882488557995 1.2942597418195239 1.14730862211315 0.97731561260268 0.8342818373545394 0.7265644333128984 0.6407267022911609 0.5522640272166537 0.4329390720549415 0.2674962760196219 0.06354191610833902 -0.15637547553299094 -0.35771975377021487 -0.502630381308242 -0.557594522393088 -0.5147121952732102 -0.3889004229592278 -0.20398691899909685 0.007095996521453962 0.22270780599582957 0.4351951824581723 0.6328776224249417 0.8015573506119996 0.9279399063891345 0.9973799206670576 1.0046109494959163 0.9544911684431043 0.8499948432087202 0.692997311714752 0.49067776190633505 0.2546339534538899; 0.1003870383988707 0.19664029039180816 0.2883102824093729 0.3796737052043088 0.4785486942144629 0.5923586882248825 0.7159938452929154 0.8208450002961849 0.8696080648128603 0.8437925723125229 0.7615326777534519 0.6795650858392027 0.6579500711336882 0.7165060337942878 0.8361587896559556 0.9811187600449612 1.103758246124721 1.1529876917635384 1.1017331076778085 0.9781282746658063 0.8521203405733455 0.7683519995035463 0.723086395311844 0.6937513144391481 0.6464140586061586 0.5483026465667512 0.3880015836311255 0.1742916267559958 -0.06579776647363471 -0.2934718861077369 -0.4699875446190872 -0.5638278490322752 -0.5610666082689584 -0.46791880125558005 -0.2994277030164983 -0.0833495175925297 0.15120673987222077 0.38766745501283023 0.6091465242158 0.7970753719629646 0.9377289736513393 1.0169401463187238 1.0275516410263785 0.9750014441237894 0.8657617408895784 0.7038121521178045 0.4969338940033937 0.25728199276587255; 0.11711317115555989 0.2259830907408012 0.32368252556792254 0.41507655464999443 0.5126811518466466 0.6313356144830028 0.770294262899344 0.8969858836825079 0.9631030955004822 0.9362908835478433 0.8250161244760946 0.6873077118330191 0.5919376113389382 0.5716936289465818 0.6238069388014627 0.7260870558403044 0.8365776901947249 0.903595122648466 0.8946906277520049 0.8332376676368034 0.7803607031694613 0.766179424560556 0.7805864586160471 0.7924012656117126 0.7578212194247086 0.6496996016376589 0.4690793359545437 0.23346751841988206 -0.029885157245068777 -0.2777886714656149 -0.46898129037186786 -0.5819435681749712 -0.6087185253346701 -0.5480547108221883 -0.4031781488871816 -0.19486106918713056 0.046828577976454414 0.30120775799697197 0.545591207803891 0.7547726538208153 0.9129543142570604 1.0058439640503238 1.02375146987609 0.9720367923335966 0.8609795827669742 0.6977385892447012 0.4912448354297121 0.2538427138955741; 0.126105433660789 0.23850508763775866 0.3303302643831049 0.406903613008823 0.4864387647683196 0.5920982150449855 0.7299748133537489 0.869043121985837 0.9565295480742741 0.9522384001961787 0.8546525198028929 0.7075139461340706 0.5687794909563496 0.48013138752483897 0.4622435660675291 0.508948276729706 0.5875554367978337 0.6582519118779826 0.6933137108314971 0.7090907782296297 0.7446763045630215 0.8105049203573272 0.8854553835896555 0.9261920759847515 0.879285387858829 0.7369527455444307 0.5114924145237358 0.24089733212422937 -0.047823975334373436 -0.32182155540285945 -0.5313227555340101 -0.6534574900707077 -0.6919990326045834 -0.650294852497981 -0.5218404999425446 -0.31775636112073613 -0.07060041348128025 0.19578738022729542 0.45817199182055773 0.6866839790690207 0.8621702858556199 0.970217641415328 0.998653504559233 0.9509915375199259 0.840539330721325 0.6786244491315699 0.4760874429558591 0.24540318114019968; 0.125256246235757 0.23140238561779888 0.30729112919099955 0.3577498803335628 0.4043559580285502 0.4760985579967929 0.5872965461776608 0.7155376458378367 0.8136716932823476 0.843244516426815 0.7952713168583992 0.6884976788336737 0.5533869321201623 0.4304627144646591 0.35834652879363854 0.34486081164729626 0.37397195903007713 0.42611694920007004 0.48836529901820774 0.5773323071516046 0.7072359632000881 0.8612088560200294 0.9968992261827874 1.055450803710014 0.9851461116947207 0.790204717643887 0.5027372585720818 0.18809123627932844 -0.11895831031989507 -0.4119434642764208 -0.6468151396330588 -0.7826292095889104 -0.8243492301063925 -0.7875832183606876 -0.6639186059273995 -0.4542725288230422 -0.19273445604842782 0.08923456447827698 0.36744533348399133 0.6120011398250037 0.8022926033766257 0.9239332263842812 0.9634997597750331 0.9223024564527346 0.8145093480202735 0.6549257755698353 0.4573118571482667 0.2349158222433197; 0.11452916904090542 0.20711855655059785 0.26374137885734994 0.28665290156406137 0.29487391863542556 0.3176388275562343 0.37777116655675913 0.466626040554981 0.5507591590872419 0.6032907708511971 0.6138815973927698 0.5762717697943507 0.48695598508658444 0.37096928930623824 0.2702903846432683 0.20484058782053405 0.18050633336899363 0.20047391673736245 0.27406810101255624 0.42222257772762767 0.6366541345800878 0.8708032993430062 1.0624064654935397 1.1357236009617588 1.043990649597272 0.7855168508207417 0.4269918285926347 0.06502113115130338 -0.25508757499468365 -0.5465959114404593 -0.7934160946107923 -0.9442698703111466 -0.9903055590536178 -0.9542184820170334 -0.8286113947132874 -0.6045798731812287 -0.3176014602285261 -0.009817392424300134 0.2892795324825232 0.5497737329880589 0.7522245158941946 0.8843658856964722 0.9334173450726673 0.8991524291832206 0.7950366282439771 0.6375709911157227 0.4432220740816185 0.2268147012801911; 0.0954959786151672 0.1712546427994465 0.21312013226206372 0.21886119421958736 0.1985825553116184 0.17460991495753636 0.17456816812006098 0.20373105807152422 0.24637739956823215 0.2929583102459371 0.33887744680252974 0.36504048036213294 0.34044269716369596 0.26043709491997746 0.1555870783052216 0.055533581042925016 -0.011896413327020459 -0.021554630746024775 0.05275323262016902 0.23112583482741617 0.4918379598636915 0.7789763757111247 1.0176709344616146 1.1121528677392745 1.0162115960684814 0.7241392418804473 0.31421246376367395 -0.08271129286638561 -0.41410386159317253 -0.6962609879899918 -0.9330758978882634 -1.0892958670831983 -1.1449114859456389 -1.1155356371804968 -0.9916751314743549 -0.7555875928988297 -0.44097315975333534 -0.10038083770023924 0.2275970710321696 0.5096535756491382 0.7258538185585472 0.866439956878365 0.9221333137227093 0.8936700042487019 0.7931313239445648 0.6365682799321909 0.44195687472588036 0.2257941328039998; 0.07026000747937208 0.12826692207689014 0.16339866579331094 0.16896026675512868 0.14250739827125128 0.09329821533178603 0.04617898844682743 0.015459165798885628 0.0018380668728279594 0.012824306926857364 0.055525916329423775 0.11299581813990978 0.13863244551831433 0.09746673026224147 0.0027034722715678027 -0.11161503008655431 -0.20077767917879466 -0.22905774853061234 -0.1701511404685366 -0.00936487475606339 0.24126681665456473 0.5433720340601448 0.8115438088805148 0.9389982445826685 0.871586278837573 0.6069414074981895 0.21104948176068417 -0.18540324150306028 -0.517419344154204 -0.787299611033412 -1.0030069724478712 -1.1516288434747655 -1.2189907570719476 -1.208577061946629 -1.0994509479647743 -0.866107837090595 -0.5390364074504645 -0.17459625429478062 0.1810463112260952 0.4885572958739258 0.7239667084031931 0.8757662808938244 0.9371098665559678 0.9123448999556514 0.8140133633425991 0.6565899145209176 0.45751332296839037 0.23422222435860304; 0.0395345440119671 0.07738621013009522 0.10977664591062798 0.12840626931473006 0.11942124860253597 0.07605634357349085 0.011660214346692744 -0.05894558144968165 -0.12365342052789534 -0.16123576917498628 -0.1526205672662187 -0.10538339091135318 -0.06626392604624119 -0.08419687619874879 -0.161499218990277 -0.2680567288248469 -0.35891984039377584 -0.40493960879946256 -0.38790394212229373 -0.2910494548301905 -0.09767522158170899 0.17986735621745617 0.45991301791269507 0.6309085817790692 0.6246714274443307 0.4452299300986511 0.1325478380591925 -0.21618765729028247 -0.5337922144511428 -0.7903294099719645 -0.9811891103390625 -1.111785443034118 -1.1821052049914986 -1.1887816769902773 -1.100893713888255 -0.8879915057206587 -0.5734954085126648 -0.2082712441477596 0.15954532818874087 0.48520262236182915 0.7395509194372263 0.9058390892269481 0.9751575557143943 0.95390834097376 0.8561613974080611 0.6953766698911282 0.4877593243888226 0.25089932630972617; 0.0032702669668071413 0.014349066464400359 0.03797637569288038 0.0695778135961641 0.09058141418440471 0.08028721163711518 0.03456087246185639 -0.040217754254410966 -0.12950414551393408 -0.2068324106550282 -0.2455207635574993 -0.2412616303826065 -0.22662610353310414 -0.24353754776381004 -0.3033436548752216 -0.38975764381166156 -0.4729301982157647 -0.5396938077042043 -0.5805035447658263 -0.5642960978409025 -0.44788010812200646 -0.22304388207025364 0.05500607726411767 0.28153830605310404 0.363251672379064 0.2965989473498929 0.1013923431257935 -0.1638208812632969 -0.44152574011986784 -0.683574428041477 -0.8624013080064832 -0.9817546127316263 -1.0506191383341055 -1.064746009139752 -0.9916004865666833 -0.8054934111916727 -0.523572471124537 -0.18188950760075254 0.176846595256202 0.5055503619743805 0.7707288061036578 0.9491376264006032 1.0269763815699586 1.009776839601775 0.9117785156923272 0.7455350589775128 0.5262479747448463 0.27190042906719714; -0.03802041904102115 -0.0627861220201602 -0.06181460814293468 -0.031401864267802095 0.014417758764001334 0.04904173611692326 0.05166439681887334 0.013878548428958061 -0.05899204760348863 -0.1474612781221193 -0.2257612613468195 -0.2791991534824294 -0.31565352159188964 -0.35433733683896046 -0.4072444128619555 -0.4736004678519405 -0.543990042840812 -0.6223293185071456 -0.7078474851211295 -0.7588165261525742 -0.716817741725735 -0.553016343026878 -0.27422268997014465 0.005854117698188248 0.17644367478378056 0.22071732244770326 0.14432405025075606 -0.022106266282819594 -0.2379529034424976 -0.45437724433975013 -0.6286555394205484 -0.7493424337815046 -0.8229955028601089 -0.8449683459571312 -0.7867807144878991 -0.6325066819608406 -0.3947415777970127 -0.09317498258385594 0.23826934692482524 0.5534856025626681 0.8172744268771559 1.0009455482009622 1.0845427778699972 1.070248435211096 0.9706040486771895 0.7971396983278946 0.5648600396134025 0.2926469982137596; -0.082374407764183 -0.1504822355308787 -0.18691743645650727 -0.1781293329885944 -0.12687626546420053 -0.05547987687404445 0.0064619633581285306 0.03532305105455666 0.01801732795350803 -0.04584096311750106 -0.14136417589496875 -0.24638332040121758 -0.34217670760732777 -0.4187501714627014 -0.478908798306556 -0.5309985869793263 -0.5823889347157727 -0.6510255895071763 -0.7477336340360601 -0.8295094694914238 -0.834387158915065 -0.7217485259199571 -0.4454200078802153 -0.1377237640841543 0.089194518185467 0.21797734704935515 0.23533220543891165 0.15726342096751017 0.010307413243333199 -0.16627996979719717 -0.3254950644818805 -0.4415002429269256 -0.515862183484204 -0.5439255092022153 -0.5044348207425026 -0.38936424474222736 -0.20386433036749216 0.04747414005331997 0.3390309010943746 0.6281681129114224 0.8790983958912936 1.0583082100067844 1.1413816636524794 1.1267489196944231 1.0234936730890631 0.8417810632266232 0.5971209835726955 0.30954870660553074; -0.12634568382523903 -0.24089030328379338 -0.32454632837328473 -0.35524935820876796 -0.32252503286127215 -0.2372543135772968 -0.12759286722309737 -0.02798243700574173 0.026960722127341515 0.01109292918482177 -0.07796346560781518 -0.21600088348801955 -0.3621195558508521 -0.4771394929535099 -0.5481495099206359 -0.5858346340006695 -0.607330558594228 -0.6396490070165048 -0.7049750350822566 -0.7702747261550088 -0.7728579560017718 -0.6872403771204516 -0.4509906429200778 -0.16234488689709053 0.08061311515779325 0.2535818247251899 0.32712494373977896 0.30704114728461346 0.21864884334802237 0.08986601333836332 -0.037641196799146036 -0.13321461385319538 -0.1930351167807005 -0.21517720721470393 -0.19006087443157768 -0.11354788987899467 0.021066224641410055 0.2209833189084123 0.46702865410504596 0.7223977987234728 0.9517623470712242 1.1176116218300818 1.1928870337113444 1.173056081541814 1.0635440055566665 0.8732679292423149 0.6186406305318962 0.3204529268368251; -0.16636288188956483 -0.3255206630395822 -0.4591655221430267 -0.5394280828266975 -0.5439107130039634 -0.4703086952934908 -0.34030345537806894 -0.19406629274028708 -0.0818702493370201 -0.04989111423127593 -0.1184046176944761 -0.26827622940642554 -0.44404003690048727 -0.5824318309596695 -0.6556767878058889 -0.6750336885781206 -0.6583297073315553 -0.627080600701252 -0.6118311683408336 -0.6107830120089897 -0.5822049319105619 -0.5078223138742345 -0.3423790335125883 -0.11248348343552887 0.11212327422782241 0.29626809567536405 0.3963519547883603 0.41108666106222463 0.36461118391179714 0.27751041817875255 0.18377814364292352 0.11583809224277178 0.08149463222847941 0.07463609790931805 0.09416545825395481 0.1444319739030899 0.24212406245303014 0.40029612121672764 0.6039851758659143 0.8241035778690777 1.0270721605989024 1.1726448207487237 1.2334609297756858 1.2034345094081746 1.0849277871279606 0.8865668611818666 0.625902843463064 0.32361039428392036; -0.20023905823068203 -0.39876574453801394 -0.5792388660948188 -0.7104716556538786 -0.7616160745037364 -0.719999148846869 -0.600383139180371 -0.4442302302835887 -0.30899221180592623 -0.2493628961611407 -0.2961174375323022 -0.44002791844076483 -0.6243139514022958 -0.7708738770231857 -0.8368104259408365 -0.8317857458895555 -0.773654603757554 -0.6698566340793756 -0.546946788073082 -0.4396143096013433 -0.3566138885529919 -0.28873571743398785 -0.19346733340680994 -0.03978323633889856 0.14718755030746405 0.32791685402038384 0.44803465744827414 0.492721139584093 0.4738164987027322 0.412668670286208 0.33988627294152046 0.28942073030016213 0.2729921377302224 0.2811076001649959 0.3032331762020172 0.3436883382656386 0.4246994936679429 0.5570838148665238 0.728696896559049 0.9188575576316372 1.0947101538679391 1.2154622417917584 1.2566999480738705 1.212169765497118 1.082576442376346 0.8777251090276241 0.6162748010987 0.3176867274371309; -0.22752968865831974 -0.4587122035440809 -0.6793464598040331 -0.8568400599385989 -0.9563032815871894 -0.9594034995899373 -0.875453909855924 -0.742416056365689 -0.6161889534371244 -0.5517088688380039 -0.5834688733246078 -0.7104414398753478 -0.8849303567308031 -1.0294028268862032 -1.0886905313048023 -1.062217864645089 -0.967436410489347 -0.8022016125587742 -0.5834881052363369 -0.36692910577547183 -0.2110570921951196 -0.131520056685441 -0.08003254349369919 0.007611015342791452 0.15440393418007248 0.32888178947139163 0.47325864542262597 0.5486298468139078 0.5491785135689147 0.5011854079646415 0.43844651969326826 0.39219932659563594 0.37845016060238096 0.3925911825177085 0.42053767562339484 0.4667462670688271 0.5494524558237964 0.6718637803993961 0.8248304591653794 0.9928732440440177 1.1430872533481324 1.2375546375532709 1.2566993531793291 1.1950801616707682 1.0540220529542548 0.8458597268180701 0.5897682983899983 0.30283499734085023; -0.24866384125671173 -0.5048682820964471 -0.7569120373480962 -0.9730523088276436 -1.1180362734169667 -1.1718676550484697 -1.1397904428949728 -1.052152187910391 -0.9559565007100165 -0.9005357129591315 -0.9205990260160994 -1.0229216912446848 -1.1754701570807338 -1.3125481174965314 -1.3706087111810343 -1.3315300197744797 -1.2135474943818938 -1.0162887270099734 -0.7447585105384089 -0.45867598351903316 -0.24001710934287662 -0.13001172282025145 -0.08376223035171688 -0.0261023574777292 0.09305495774777525 0.25718282417223165 0.41624930442232977 0.5112270323700108 0.5295501747239879 0.4974267200414659 0.4462514086272235 0.40570167601975277 0.39721578476013486 0.4178037443684104 0.4558594318334459 0.5174799462783557 0.6120712848228888 0.7360647974505766 0.8824361948989551 1.036017936402031 1.162969714295583 1.2314769898055775 1.228575414979208 1.1505061039577975 1.0007066012136883 0.7940817097496444 0.5493024847826564 0.2806969915712188; -0.2649481751386248 -0.5382357879497176 -0.8109660302303784 -1.0555219086448695 -1.239808937527272 -1.3441838179855417 -1.3699266345729408 -1.3379268582835513 -1.2822858983111296 -1.2419612161788935 -1.2493793684817722 -1.3190845353515472 -1.4385077552497367 -1.5643905176669313 -1.633348179851385 -1.6029129060099567 -1.4843604647471191 -1.292131164701779 -1.024232696152968 -0.7262361776283028 -0.47875971863889816 -0.3347674572756933 -0.26668949536460196 -0.20402977101635675 -0.09872545805213116 0.04914414417377276 0.19882284465297728 0.29748279058633614 0.3369055132914256 0.337790709867074 0.323422867783371 0.31541771547071884 0.3304560914263305 0.3683666406930807 0.4253056174402507 0.5080853521467101 0.6198493171279557 0.7538986845432466 0.9015200777758305 1.0429064416247287 1.1469205744986024 1.1922764456688635 1.171090507733476 1.0803570463239207 0.9268470531766626 0.7276061741475901 0.4994177911972959 0.25387036740471836; -0.2777316483767485 -0.5610354483871469 -0.8430348952995579 -1.102720114181941 -1.3149977648443507 -1.4624243730402717 -1.541780825499937 -1.5640418845983772 -1.5500749958928277 -1.5265543196221938 -1.5215623624805168 -1.5561112893347249 -1.6340529789916463 -1.7373821439774053 -1.8212869062311021 -1.8292588798359468 -1.7481363854352703 -1.6003294520434588 -1.388109821655481 -1.1331857933988658 -0.8967645232122236 -0.7287985735152963 -0.6252810842099686 -0.5381100242823071 -0.4365086301409837 -0.311963754702862 -0.18669670778136155 -0.08451746151660722 -0.009197802622012875 0.04766154580118715 0.09276873493830547 0.1360954124941214 0.19146314942994594 0.2609427717228454 0.34614548147830715 0.4563854381580207 0.590579609964687 0.7382136903994947 0.8857777276739761 1.0121444069035344 1.09494902657221 1.121606919984251 1.0861241512873565 0.9879087163198857 0.8376251802432015 0.6519938611718064 0.4445256019065788 0.224783960616863; -0.28573512323576994 -0.5724320160732796 -0.8538975989758654 -1.1156706271800014 -1.3411765631667483 -1.517255971969747 -1.6375373721307924 -1.704377859039727 -1.7270743840103995 -1.720303448960127 -1.7062321314215363 -1.7106882730373996 -1.7484303485166162 -1.8178361033046986 -1.9014680042158567 -1.9561735544903731 -1.9443236773968264 -1.8722384092014408 -1.7472974344815708 -1.5727614933716345 -1.3823715110969599 -1.2149113035856536 -1.082879227385759 -0.9671267447219233 -0.8526380308565471 -0.7345394210507417 -0.6156322963044453 -0.49747482915033253 -0.381627999481323 -0.2722013844792221 -0.17529721289714384 -0.08403732901364719 0.014565373154063628 0.12254644121751565 0.2460722563385349 0.3896069844695625 0.5452508546905888 0.7003136074977074 0.8406917921727933 0.9499802376236622 1.0137313762926683 1.0241446537584042 0.977791152134623 0.8781408919022242 0.7378470698802772 0.5707925330100735 0.38715640842037974 0.19496399276568863; -0.28504847955289636 -0.5673880779849094 -0.8403176521711674 -1.0937443730279575 -1.3186379179272816 -1.5075698539956435 -1.6532796533887741 -1.7515002030317233 -1.8032296759249928 -1.8141562907385445 -1.7987075848960643 -1.7827205492077498 -1.7892687870234694 -1.8256530710026249 -1.891790102828613 -1.9711984460575067 -2.024608807110656 -2.032688656054962 -1.9961976147475988 -1.9111920836091583 -1.7893448673518906 -1.6542382931415398 -1.5221174166182898 -1.3937363792810413 -1.2641903214170582 -1.1330145624524777 -0.9968854798799909 -0.853748592971923 -0.7068999677990584 -0.5653585739708725 -0.4337602553172001 -0.30121248312539134 -0.15928592347354417 -0.007318625869567721 0.15694661395492693 0.3283863556808484 0.49560517129994675 0.6475481324255629 0.7751757528308421 0.8666811054162067 0.9116903513361134 0.906638416622811 0.8525352349178345 0.75675063689762 0.6312848830755486 0.4859773470451961 0.3282856117552073 0.16484662134148073; -0.27361426316923715 -0.5419720462264199 -0.797928546374232 -1.0342195330634039 -1.2473539918630894 -1.435614115527081 -1.5931970601770808 -1.7116163784195062 -1.7857406962846814 -1.8159487250081092 -1.810392811901411 -1.788498717188154 -1.7751418154421512 -1.7855511394207981 -1.8278240892742403 -1.9049783862908936 -1.9935947069511326 -2.06085014430038 -2.092575427103428 -2.0804138054509003 -2.025610636801733 -1.9403041492528192 -1.8368700593801128 -1.7209163531992242 -1.5927937215506651 -1.4528645527379611 -1.3009596700273054 -1.1393024660104818 -0.9741632496847107 -0.8118012261440081 -0.6490372054328181 -0.476071863495531 -0.29212434740147236 -0.09956330209789675 0.09622319184445077 0.2827969670828964 0.4483574984071154 0.5889171126862708 0.7000728288402793 0.7707046919158829 0.7950959935170724 0.7755720197121955 0.7179980317149018 0.6309933541151377 0.5227155001546131 0.39975713408252334 0.2687257857803887 0.13472500129489318; -0.2515979250343533 -0.49578813833394847 -0.725416238455241 -0.9360731498880035 -1.1287561397141632 -1.3062763239340147 -1.4653114060102015 -1.5957134510882924 -1.688050850005599 -1.7391598165017232 -1.753152961113515 -1.741775068638831 -1.7242375474171192 -1.7190576695945754 -1.7384135682951571 -1.7941096442859874 -1.882983027411612 -1.977418340873741 -2.0524432636337795 -2.092380745203695 -2.0912987947307577 -2.054129872054855 -1.9883112797662978 -1.8979862223970185 -1.7845950325940807 -1.6496887937787992 -1.4960907136463408 -1.3288776246287124 -1.1544501346789164 -0.974736546339442 -0.7836319440851989 -0.577391568476602 -0.3605839971619907 -0.14076106244769213 0.06852216946270748 0.2523841990950951 0.4057934619049482 0.5303440962300695 0.6195568983626921 0.665050709143761 0.6689996893382281 0.6394651270337365 0.584442057486685 0.5092538507478587 0.4176656009330615 0.31600324510088756 0.21108241401236905 0.10573573650313867; -0.21920955470765358 -0.4297292710074709 -0.6251502671927354 -0.8041033859902469 -0.9708605492131615 -1.1309243201993155 -1.283662745006427 -1.4197132768099545 -1.5272037015305069 -1.5997445538130413 -1.638166642479112 -1.6481835212718285 -1.6414213055176823 -1.6347844911884175 -1.641847932652568 -1.6734244228365929 -1.7378829072158848 -1.8247161812365686 -1.9115111836043661 -1.9797889756918192 -2.0176803670948837 -2.0230129517241116 -1.9963968966069638 -1.9382015339090852 -1.8482486370425033 -1.7288888998070246 -1.583821259088843 -1.4184254395791647 -1.2377425490050602 -1.0427553723256282 -0.8303704028705365 -0.6033700118875563 -0.37102283239205547 -0.14666538571484333 0.05467022020815147 0.22478140424123338 0.3637136054881354 0.46872155535625554 0.5320300038509831 0.5533346100223454 0.5423229604502999 0.5100055540698419 0.461897728560019 0.39826142716884116 0.3217754910446095 0.24006334718321964 0.15897596328872934 0.07929376285253886; -0.17656768435448675 -0.3456146587348594 -0.5028325792731442 -0.6489134832510879 -0.7888782732103744 -0.9285284104646917 -1.0694234414278092 -1.2053340891993227 -1.3243008016135474 -1.4167456505425966 -1.4801231948967195 -1.5155495285952019 -1.5279678559633352 -1.5300378203485006 -1.5363848894377663 -1.555866478846574 -1.5957529917558184 -1.6569806264687135 -1.7276807968962427 -1.7944072028570013 -1.846277275044792 -1.8773829340597263 -1.8832530913536174 -1.8588240927720954 -1.8001000941341365 -1.7064247668142027 -1.5798209070827778 -1.4239630940596593 -1.2433301132542565 -1.041178690094087 -0.8209201152218065 -0.5912139976411729 -0.3642766943262201 -0.1533707289421981 0.03254782954989338 0.18983996808200107 0.31410613313925073 0.3978237676056098 0.43821310796503776 0.44335180439276756 0.4269304108025723 0.3980729840355885 0.3574223518217122 0.30372615060213415 0.24120954314794385 0.1770145870670936 0.11528119769127089 0.05664183271677165; -0.12604233102338483 -0.248700203471324 -0.3672110187684127 -0.48336778880446907 -0.5999337066955381 -0.7199000647797837 -0.8458228765743019 -0.97621151776203 -1.1018399156761567 -1.2109586999555357 -1.297224821923503 -1.3579983941274922 -1.3929794124000492 -1.4092787036497223 -1.4202758773248338 -1.4364343339068177 -1.4626547189398533 -1.5007944771882988 -1.5468040892626465 -1.5941007669097738 -1.6371919303018188 -1.671114601736596 -1.6911301648593755 -1.6898836649990494 -1.6600158698203444 -1.5963411761970896 -1.49732235583601 -1.3635692436309288 -1.1985663454405586 -1.0074431973490519 -0.7986323339788434 -0.582977065505654 -0.3722271165663359 -0.1765773051899184 -0.0024673843941558113 0.14318841166564833 0.2515000987305498 0.31700272019298414 0.3439550598698426 0.34500091189329823 0.3321706140363014 0.30952166204877757 0.27577338819173014 0.2315375453256844 0.1812845022217478 0.13034132365021348 0.08253988843357733 0.03951673644669574; -0.07400587408503413 -0.149819690542476 -0.23062861801839438 -0.3194330879348416 -0.41639695773431334 -0.5201374815673682 -0.6314474290755177 -0.751913505673991 -0.877561746451093 -0.9981237567774147 -1.1038877731463672 -1.188234194544257 -1.246778954202168 -1.2814417249514574 -1.301766450357817 -1.318382228434276 -1.3371299652442021 -1.3589405976423266 -1.382925997129001 -1.4079966198459521 -1.4339062619812393 -1.4588463190694254 -1.4792577340272435 -1.4887553225936654 -1.478759168430078 -1.4418085748599374 -1.3732799981184212 -1.2708266248963216 -1.1351594389422635 -0.9707725488676173 -0.7862611341641913 -0.5913836187969751 -0.39694895730546387 -0.21314812801890895 -0.049949241758827326 0.08231794975327962 0.17620943775732323 0.23166512220830812 0.2574443162917144 0.2648390637503988 0.26014541517510803 0.2444964990227117 0.2182640312226401 0.1833093026913351 0.14280414955837883 0.10115740344105761 0.06278283938870387 0.02958434381567372; -0.028585474897674826 -0.06263346993906918 -0.10815307122726575 -0.16966127373497228 -0.24721058083263228 -0.3370296143050837 -0.43641503328232967 -0.5458974422682636 -0.66527721574636 -0.7891126762642549 -0.9076438327859601 -1.0106400026440696 -1.0905008287848457 -1.1453582413411358 -1.1804012961096624 -1.2036433124007755 -1.2208163525192415 -1.2333902885176826 -1.241928756348235 -1.2492291988732984 -1.2591400215909732 -1.2736394956148724 -1.2907608016771026 -1.3052987223261405 -1.308880538211663 -1.2934689886029132 -1.252532278271463 -1.1811876672547548 -1.0770808080335084 -0.9428949008428584 -0.7849584097239106 -0.6115397456700415 -0.4331397887186654 -0.2621447855193414 -0.1115850163489491 0.008761825437077878 0.09519118237278167 0.15143441621724088 0.18559156618659095 0.2033649889431852 0.2067726507835 0.1976407649080307 0.17856114216029473 0.15188940166708811 0.11997228783001848 0.08624000526711344 0.05440625395471765 0.0260243663246901; 0.005227606754107228 0.0036922282091194788 -0.011649602572230171 -0.04661049704258039 -0.10299806395785799 -0.17746154278601706 -0.2652732188933128 -0.36452288340520794 -0.4752382216375172 -0.5954286808440258 -0.7178353018656326 -0.8308930505810882 -0.9249211842600488 -0.9962359511027771 -1.0466629000756698 -1.08099681686003 -1.1033923575071756 -1.1154514969172402 -1.11861603414604 -1.1174760346830326 -1.118527425898087 -1.126544904993686 -1.1417565818894522 -1.1600231585650957 -1.1739580630061812 -1.175082921167418 -1.1553247828248518 -1.1077227632483575 -1.0286581048905408 -0.9198760797065442 -0.786270435187782 -0.6346861522839741 -0.47465793447653154 -0.31904985924102336 -0.18035495891792833 -0.0663577749619666 0.02107437717145434 0.0851540935231133 0.12960163285404502 0.15555112246395766 0.1642387819294532 0.1593260867597421 0.14555385827696846 0.12648489806654717 0.10363722839588921 0.07797267742724236 0.05132157517180626 0.025285885044968807; 0.02758992501660755 0.04820916008614449 0.05513740853235976 0.04248301461972258 0.007208165178572009 -0.04929329261966967 -0.12328996006456935 -0.21191499731493835 -0.3136551441575122 -0.4265331344658432 -0.545052696818883 -0.6589686891645812 -0.7582076647501362 -0.8383310500530055 -0.8995041513179692 -0.9439301479229256 -0.9741725753214477 -0.9915330065619307 -0.9977552295418016 -0.9977101351818886 -0.9987065415345932 -1.006455230915621 -1.022519190670267 -1.044203162326917 -1.0652866135641175 -1.077138834517065 -1.0704953542497948 -1.0375541010004456 -0.9753655694861914 -0.8863057993418125 -0.774740734030772 -0.6455570554176892 -0.5061935237358677 -0.367361932984568 -0.23919337158146514 -0.12830281821065243 -0.03726339636058258 0.033804129843938414 0.0844719209532788 0.11422677407317712 0.12497584079315827 0.12201335855917235 0.11221075173123664 0.10023955981719018 0.08629312049150265 0.06859530461496537 0.047193025440550346 0.023879874069589063; 0.04064133272178401 0.0741290140916542 0.09452872086220127 0.09738227619696181 0.08024828107480687 0.04259853367176433 -0.015264315375893525 -0.09197679626961007 -0.18433326072084877 -0.2878837724120293 -0.39679860778711856 -0.5028927972218908 -0.5978574057311731 -0.6777069656342851 -0.7423425326739003 -0.7927249654942623 -0.8300494165880185 -0.8550947662431212 -0.8691815009230935 -0.8766088014286827 -0.8839839136754197 -0.8962866172083362 -0.9149996941346411 -0.938691772962998 -0.9624954808407366 -0.9782919098715841 -0.9767612828006187 -0.9510571597406294 -0.8999335122963403 -0.8268372549933337 -0.7353378633577212 -0.6281492250121382 -0.5101380486599693 -0.38914840855516414 -0.2732235767465085 -0.16837868915242887 -0.07838227614859518 -0.0061804888655291575 0.04542616316065822 0.07534018265241552 0.08598967435770072 0.08400748919632176 0.07746937900824657 0.07122912838996066 0.06462304245313889 0.05434402616978545 0.03912958738321443 0.020364939696053283; 0.04537031068596887 0.08390078104653949 0.11048134396287673 0.12217188510396215 0.11786850526218628 0.09660594661208477 0.05577987737206913 -0.006436059975115702 -0.08715506409313566 -0.1796112740526306 -0.27603602577156333 -0.36859469959204405 -0.4510595562225228 -0.5212157306826447 -0.5798934760501552 -0.6285305021589863 -0.6680728866022259 -0.6988856962124675 -0.7215376278175752 -0.7389152811691723 -0.7556659628209959 -0.7747807622682331 -0.7968258207561327 -0.8208634157231437 -0.8433233369344613 -0.8573526758781642 -0.855305635178686 -0.8327048743477607 -0.7901409812854832 -0.7312899113239223 -0.6584562696887533 -0.5725009089786577 -0.47601857949748666 -0.3747542185723716 -0.27540380437169054 -0.1831737006914348 -0.10182074038169707 -0.03524931780457811 0.012540550014051867 0.039791173169141346 0.0491801341266471 0.0478719639482481 0.04376937918068699 0.04137278281296644 0.04005177578098609 0.03624356771545867 0.027726750028452478 0.015008146964804231; 0.042091267272438435 0.07864676299952121 0.10580842376767023 0.1212954010324634 0.12421351152423205 0.11401777054636347 0.08802818887779838 0.04273809892371532 -0.021246255587601598 -0.09797677408427424 -0.17899112701936187 -0.255875207437407 -0.32271007835089344 -0.3780605398712999 -0.4237948382407616 -0.4623654075757671 -0.4955710489375142 -0.524436118124976 -0.5495816780494147 -0.5725399842216614 -0.5956387213645987 -0.6194382269275892 -0.6431581559120277 -0.6657105778206026 -0.6844721135063421 -0.6943828472239666 -0.6905338378609684 -0.6711252624909896 -0.6379753032934838 -0.5940062197114179 -0.5401222483147091 -0.4758298041911813 -0.4021676135871992 -0.323410520358649 -0.2451012119674992 -0.17149865787532687 -0.10552023042256466 -0.050550082937106826 -0.010763189440408176 0.01166396417256468 0.01931001743329589 0.018614628221435114 0.0161758553099998 0.01590362713302265 0.017669112678716098 0.018506915906971528 0.015765868557967692 0.009090670544440073; 0.03191274369887864 0.06024229732903089 0.08266179510658193 0.09739451923547268 0.10301028530262771 0.09880740267014898 0.08348180822626741 0.05466645617875523 0.011662549791275387 -0.04246980388528156 -0.10174665980583089 -0.1592156586428931 -0.20900286781603455 -0.24883122768568136 -0.2801118990414922 -0.3053668896968462 -0.3268874241345493 -0.3465689806431971 -0.3657039940266968 -0.3854330683167203 -0.40654986175185626 -0.4281087681427085 -0.4484534093978045 -0.46639525921017216 -0.48001652440960174 -0.48605485276008326 -0.4820959007145761 -0.46827574033131336 -0.446494432718961 -0.4184636829671622 -0.38410774603212827 -0.34231293339914587 -0.29311945541006007 -0.23934621855662042 -0.18523815284450662 -0.13410171514071156 -0.08788732753822118 -0.04889528830238959 -0.02046790772334356 -0.004406304621426101 0.0012699544266397181 0.0011642802847262794 -0.00016255621985892757 0.00034467631107543964 0.0030645339089468634 0.005927368724378735 0.0065752550136423185 0.00425605204167159; 0.017022036052498933 0.03243675507683257 0.045154811995183154 0.05423255389989477 0.05854035223124336 0.05723066787986223 0.04984523003096947 0.035760436237211576 0.01458080755390123 -0.012783904618226642 -0.043863215826521924 -0.07515039971324969 -0.10286204900248495 -0.12481501758545883 -0.14126027997697405 -0.15369388021872687 -0.16369148546326862 -0.17276866183275513 -0.1821582009520169 -0.1927369951385338 -0.20467625615656418 -0.21705806052301926 -0.22862491214159658 -0.2385435112448802 -0.24572126215589568 -0.24861238597896373 -0.24633606095906468 -0.2394678426437066 -0.22916786072865078 -0.2161191512817835 -0.19996252473812776 -0.179826031096638 -0.15546206449981717 -0.12821518921483316 -0.10045724839569158 -0.07414240234491863 -0.05032508054401927 -0.030116586184948897 -0.01530139779539877 -0.006822033688186716 -0.003675287194829899 -0.0035242337188272537 -0.004090489589853737 -0.003664813602973828 -0.0017804847560341 0.0004736729875959821 0.0017114455626505525 0.001379307673420638], hyperparams = 95.61750867171621), (params = [0.005123277443138685 0.007041833324956083 0.00462785953632739 -0.0022034712220318294 -0.013852922315443291 -0.030551311309884987 -0.051926733041947004 -0.07665430708792392 -0.10270349148162103 -0.12787090383121896 -0.1503589345737174 -0.1688764563397034 -0.18296963595698987 -0.19275663930992626 -0.19926834403555224 -0.205047962739104 -0.21237861709759728 -0.22227386641185667 -0.23333065638351658 -0.24226305075614732 -0.24663536505057146 -0.24538766191389857 -0.23852511490434616 -0.22744100644270251 -0.2153573581421105 -0.20476852539099916 -0.1956410284483593 -0.18613227719983452 -0.17435949119470784 -0.15944146102737894 -0.14209379037808803 -0.1238863961519179 -0.10619477690158388 -0.09014263591857825 -0.07749657538765066 -0.07001147791874592 -0.06863254912753095 -0.07245319282513138 -0.07903706507034193 -0.08575500093768451 -0.09127828429995742 -0.095629444172119 -0.09924890460332919 -0.1012154542001495 -0.0986444415844289 -0.08786461894933888 -0.06671232259245641 -0.03617902303062271; 0.013143197381536624 0.020350669313317964 0.018594826928910187 0.006354397820233398 -0.017768217097388427 -0.053521349720671615 -0.09824762828898717 -0.14793215871426035 -0.1981471875980001 -0.24536975664757332 -0.2876622289359342 -0.32355825021602136 -0.3515501410634739 -0.37137007707254077 -0.3859379644178559 -0.40051171010453046 -0.4196810001709396 -0.4444629704238842 -0.47079636157725085 -0.49165868957260034 -0.5016871237892416 -0.4990689776611992 -0.4842102807496379 -0.46030263373062624 -0.43378538209670553 -0.4095742624348722 -0.3879609240993524 -0.36582090102681797 -0.33982681607899906 -0.30857663298444243 -0.2737998275209016 -0.2387827261319979 -0.20626186917149064 -0.1783125104526289 -0.1565925490332617 -0.1426243780027627 -0.13737541644211046 -0.14009415482246804 -0.14844875489522988 -0.1597351136992702 -0.17157761516627118 -0.18226275900760058 -0.19023211114813707 -0.19272571799172078 -0.18536075920344144 -0.16317319040671857 -0.12310747742995932 -0.06654946255696048; 0.025591418920287448 0.04271971826081542 0.04549497469965896 0.030001400773587455 -0.005886480537640927 -0.06058957420889771 -0.12838275766925586 -0.20196040228401752 -0.2746400761063459 -0.3422667325038067 -0.4030219365638412 -0.45471174299279354 -0.49492930021711057 -0.5250880251337035 -0.551553964052742 -0.5818883807421326 -0.6209855319132539 -0.6673315520945133 -0.7129304715158953 -0.7469646927638589 -0.7619612528017528 -0.7562303696996902 -0.7313082358395709 -0.6926315209206417 -0.6489866517615404 -0.607115488067211 -0.5679224815097921 -0.5282301830997937 -0.484365498736198 -0.43544817705621874 -0.38417261225627464 -0.33497089146203796 -0.2916648876466803 -0.25705829724800844 -0.23179779970338749 -0.21517175048710765 -0.20643661788153275 -0.20501165952338915 -0.2100611439108135 -0.22063089688955806 -0.23523763206843126 -0.25124305051965345 -0.26384658013825013 -0.2666051263433414 -0.2534153425893812 -0.21985628550654984 -0.16403577738672942 -0.08815189196341962; 0.0421633092540315 0.07303488955090762 0.08352889633262767 0.06769185171875593 0.0234525563973217 -0.046177211736561155 -0.13330212494116508 -0.22805397755193105 -0.32229426792054744 -0.41098526348064085 -0.4904645020621237 -0.5568517358824775 -0.609690338827475 -0.6551644908307727 -0.7025846270121029 -0.7588731778966186 -0.8247552108326801 -0.8939685004382414 -0.9554763200663118 -0.9972454692727307 -1.0125395754928619 -1.0011234584025497 -0.966479484341482 -0.9154302466788335 -0.856982846758877 -0.7975406631853139 -0.7379492604703929 -0.6759680068937592 -0.6095249060067899 -0.5399282378313638 -0.4714648782105017 -0.4088236874500408 -0.35618362277562665 -0.3168929475708859 -0.2916461073351968 -0.27755563063314886 -0.2710672239770903 -0.2693997738664928 -0.27103073682829565 -0.27630345917031623 -0.2864767098648518 -0.3013455528354555 -0.3155283372573888 -0.3185749419132547 -0.3008434431592139 -0.25790571311877847 -0.1900783317154115 -0.10135075712178497; 0.06063286689247589 0.10727062984268021 0.12790114725333138 0.11508335922893326 0.06699937324865746 -0.012492294139488862 -0.11413007725058996 -0.22639895273894808 -0.3401727004884916 -0.44897042222463646 -0.5467398071788583 -0.6298998389003583 -0.7026766447457901 -0.7752746694155274 -0.8554596676875776 -0.9435936956733243 -1.0333709478340056 -1.1154497897718827 -1.17996153111437 -1.218928809784452 -1.2294266681946389 -1.2138660525111953 -1.1772804736825238 -1.1255583175463602 -1.06480149009964 -0.9974892209268771 -0.9224845274973642 -0.8379124715864267 -0.7449162609108346 -0.648656537014066 -0.5567979785076869 -0.47510208764395445 -0.4080156924351148 -0.3595371004901225 -0.33151536296083545 -0.32092123181266585 -0.3216361754030171 -0.3269140126249473 -0.3311872044692832 -0.33300084264486524 -0.3352502212545553 -0.341779225353574 -0.3505539401386064 -0.35064168822952146 -0.3292308649706733 -0.28009396486761096 -0.2045216066793124 -0.10820747684806088; 0.07837332102394105 0.14073242468113736 0.17276487324531487 0.16562168604607355 0.1172468419218738 0.03184727066843284 -0.07999762152306025 -0.2055126506665552 -0.33492513861347345 -0.46086559171182123 -0.5770179059523399 -0.6830901086107591 -0.7874534778027714 -0.89866557343088 -1.0156718952595165 -1.128771304393089 -1.2271151031891252 -1.3029716778796043 -1.3537003402605294 -1.3801448413984412 -1.3849463763449683 -1.3732179945268947 -1.3496637341664508 -1.3171797155040952 -1.2754618087819662 -1.2198792907957896 -1.1445227892890917 -1.0463711295308142 -0.929165206897269 -0.8024617822545622 -0.6782537121096182 -0.566217299488166 -0.47288390569466365 -0.4039267381323584 -0.36286171311469206 -0.348664264102509 -0.35402792658963333 -0.36866634599967374 -0.3823674463394434 -0.38846863840213336 -0.38710166986454686 -0.3834055310050169 -0.3801386906071416 -0.37119573467914524 -0.34433483719727453 -0.29093066329646927 -0.21116376999742512 -0.11114668554646207; 0.09404130141375376 0.17067446035502715 0.21395960414352763 0.2136333971284447 0.1671027031596277 0.07899883520042242 -0.039320905233579534 -0.17454953962089903 -0.3166165196367504 -0.4583689102149047 -0.5958881914826524 -0.7332523652193582 -0.8787218381549053 -1.031099768227682 -1.175889177974996 -1.2963352659327227 -1.3817420545826733 -1.4305945486846183 -1.4519918900596784 -1.458005275668985 -1.4582274267803683 -1.4593552314398677 -1.4640954297895468 -1.470628255733044 -1.4703822266574047 -1.45019293513524 -1.3967521034378627 -1.3039219130555608 -1.1756008403577354 -1.0241282801503608 -0.8649354605494818 -0.7133073180709881 -0.5809316145485756 -0.477254399850879 -0.4086564171180408 -0.37701571152961433 -0.37655923052237894 -0.3951638888228073 -0.41907711983306767 -0.4360681728237972 -0.43951503506407746 -0.4305988061780783 -0.41423564649860556 -0.39148253726758175 -0.35566614272845976 -0.2978448639906415 -0.21535996048125125 -0.11308988181382736; 0.10782533981364605 0.19715659631998286 0.25087922178278377 0.2574988411085 0.21415091198702949 0.1261146225732374 0.004444262976216984 -0.13813864214186614 -0.29195690529181495 -0.45163515072334903 -0.6177066572783072 -0.796152441341929 -0.9870058544791988 -1.1731139920807852 -1.3289296143678835 -1.4368327229514193 -1.490333857997708 -1.496183444936194 -1.4757835420182757 -1.4544156176315755 -1.449635660449754 -1.46816994720294 -1.5094148612126348 -1.5653934005013386 -1.6203056955801927 -1.6532238334994453 -1.6433142134833882 -1.5793082481646747 -1.46223371636049 -1.3033829723738395 -1.119063999542439 -0.9288253773413506 -0.7512864197359643 -0.602090950816039 -0.49275185238223157 -0.42878763120195146 -0.4080222004581453 -0.41912719088676953 -0.4462827948700003 -0.4733636218358292 -0.48709216690314394 -0.4811493211376391 -0.45733949367585236 -0.42114506724499784 -0.3737905273735178 -0.3096377814062559 -0.2235479123164522 -0.1175512873695136; 0.1207383126558213 0.222113454313086 0.2854983063196785 0.29864849702135843 0.2597908243017677 0.17478881059538676 0.053163348837696515 -0.09432105775065781 -0.26047979425075496 -0.44465778407643197 -0.651005105773888 -0.8796490935375231 -1.113935846040193 -1.320534575163797 -1.4697714926120156 -1.5493521708401095 -1.5601655818697338 -1.5164345286797036 -1.4505616472848408 -1.4007880985106187 -1.3909598880100436 -1.4268456518304924 -1.5027640266604945 -1.60416747715921 -1.7110159448360824 -1.7986230949390674 -1.8423232736616455 -1.8256289506817727 -1.7442133094032704 -1.6037090934795075 -1.4165760127233684 -1.2025257289753306 -0.9852980344496521 -0.7880550458116988 -0.6302120988438527 -0.523154392251909 -0.46900754663603356 -0.4591665336339263 -0.47755482205402416 -0.5060398976415234 -0.5279408915242668 -0.5302284447375174 -0.5073248778278606 -0.4634661641655174 -0.4054539192574992 -0.3327655122532963 -0.24011787981475244 -0.12668901740974586; 0.13357082017847452 0.24613468222879314 0.31799453620322404 0.33777381987009486 0.30566196392891776 0.22743442415637796 0.10980107741336868 -0.04108486588388265 -0.22391959032896708 -0.44336857233085913 -0.7015230344288977 -0.9844414790759455 -1.2551874561465355 -1.4681942019984704 -1.5966737365183286 -1.638364648935161 -1.6036752736402706 -1.515032402014974 -1.4137316043282473 -1.3448441213759115 -1.3335366503467045 -1.3825112105552766 -1.4800353497419558 -1.607768931913156 -1.7455198260289895 -1.8703026113810426 -1.9594999819933085 -1.9951605648969513 -1.9673990910486938 -1.872426937344031 -1.713448813015682 -1.5040448336485406 -1.2676470634466475 -1.031799513044159 -0.8251958379894099 -0.6689263226804548 -0.5717297802304369 -0.5292426627010062 -0.5264951282747918 -0.544880464140251 -0.5669179463686733 -0.5762791585759666 -0.5599229207444056 -0.515575170718901 -0.4504000312099298 -0.3681372762991888 -0.2653375116557438 -0.1401866444879994; 0.14518741743458713 0.26714660307019295 0.3462188836926916 0.37350086396568494 0.3510286319264555 0.2829831090168173 0.17104681289680296 0.014361892816288723 -0.19234647063791457 -0.45579055383111006 -0.770919111816266 -1.1062119680345734 -1.405358817765968 -1.6160426230201181 -1.7175159064442003 -1.7178782826475119 -1.6399388290875232 -1.5184001989646296 -1.400676908589048 -1.328918683184467 -1.3218059502969934 -1.3745897944995753 -1.4703897173877396 -1.5930040726303836 -1.7282494961296289 -1.8596603368699733 -1.9718684476672144 -2.050171129212809 -2.0821973295578613 -2.053780849710188 -1.9553132723111581 -1.7872191440958847 -1.565924400007084 -1.3167731880549518 -1.0729016346381104 -0.867245689442098 -0.7199738133690857 -0.6346489317105075 -0.6000493683425429 -0.5982510132032738 -0.6115880853605589 -0.6225759761634614 -0.6131118671974278 -0.572690758221462 -0.5037908257518092 -0.41172022062373836 -0.29601820144215196 -0.1561475029750622; 0.15396433023105358 0.28260628055587533 0.3674960051818706 0.4030145452801898 0.3918261625254288 0.3343624959972065 0.22616195266167213 0.059814150580849976 -0.17468465206892672 -0.48328121169501553 -0.8523662779999308 -1.2344970805504423 -1.5586746226320813 -1.7678562584268471 -1.8444948706205997 -1.8061308109451542 -1.6908283191917273 -1.5490894916256448 -1.432803538858029 -1.3739807422884753 -1.376176307652725 -1.4207123471505487 -1.4869282300579199 -1.567760619748607 -1.6617587488543017 -1.7641023501472088 -1.8702908513899397 -1.9730030881169722 -2.0606344815412743 -2.1100832061443477 -2.0971336257178863 -2.0048651613903763 -1.8357674433968934 -1.6088623913889566 -1.3539648094089516 -1.1105513488966305 -0.9125285417689919 -0.7765375406250089 -0.7005835510633618 -0.6700726943624177 -0.6672426529072458 -0.6734645275980866 -0.6670778454519909 -0.6304705460282024 -0.5592678703883548 -0.4574798815424181 -0.32768840862887416 -0.1721641525407308; 0.1581460808372101 0.2897560903129787 0.37835722073880335 0.4210854715037343 0.41953604369965797 0.3695150871922133 0.26155052451924843 0.08389278263593275 -0.17592785528155638 -0.5226169363424118 -0.9364443264562299 -1.3586663733675246 -1.7067945228648778 -1.9181440100429514 -1.977430132128548 -1.9102555707285513 -1.768257704501938 -1.618088934357874 -1.515822406470132 -1.4811281884044993 -1.4946432574073403 -1.5178446006489061 -1.5290925596116167 -1.5346446140060368 -1.549131127850643 -1.5861202943668142 -1.6561409746694058 -1.7643937632517508 -1.9005552012367388 -2.0339351848868583 -2.1226319360086983 -2.131245619486144 -2.0429582817123926 -1.8688244843936093 -1.6344795744459528 -1.3778390797945486 -1.140696076934719 -0.9538888942279767 -0.8298754453753906 -0.7630958474931261 -0.7374302889040284 -0.7324273311197101 -0.7234676846429671 -0.6870643914957406 -0.6122048940421717 -0.5005668954769773 -0.3569821337293098 -0.18664209459238576; 0.15639359412526904 0.28600917290963607 0.3741014499036298 0.4192085095771212 0.4210514232199934 0.37209114574897056 0.26071005462493163 0.07338449196225277 -0.2036177310147575 -0.5753273105746513 -1.0197609616688765 -1.4701635283021457 -1.834984267580427 -2.0503887129588843 -2.1052999397292016 -2.0284503534151312 -1.878670086412361 -1.7337658523034467 -1.6536711763031593 -1.6449281745663216 -1.6608526802571304 -1.6459165691101458 -1.5818372724017418 -1.4857500798421241 -1.3883836608317286 -1.3267603452373808 -1.3342283927143097 -1.4333323989527706 -1.6145672975549903 -1.8368783578071723 -2.0377136117469528 -2.161997628262919 -2.1740369352423823 -2.0729216929472325 -1.8825695640748554 -1.6392151285950107 -1.385148826642587 -1.1592866681421605 -0.9882745398353322 -0.8802988875643749 -0.8254356988525537 -0.8027112562836177 -0.7846488875748703 -0.7430941102452252 -0.6614787178993511 -0.5390148912788721 -0.38224096966134435 -0.19887701290074566; 0.14889588636712128 0.2704437932967104 0.35038615755278585 0.38787447008217146 0.3821592676656662 0.3254843223877602 0.2072389265502917 0.014676893538208525 -0.26661995945155453 -0.6441233858255746 -1.096914124619243 -1.5548676367904535 -1.925282417892297 -2.1503117618660044 -2.2215014502455457 -2.163789691190931 -2.034758323740184 -1.9129469217488806 -1.8557952664959796 -1.8555494196777906 -1.8468281999589324 -1.7712144465569668 -1.6148103327248131 -1.398834448665366 -1.1675937038416082 -0.9847982585355614 -0.9150791626766337 -0.9999181695723941 -1.2292286142088515 -1.5442322686097623 -1.8605910718418546 -2.1041279069495356 -2.225616843167936 -2.2113533724760654 -2.08056180502074 -1.8699258378556856 -1.6214008838599605 -1.375720312852829 -1.1682799769931356 -1.0205027585003907 -0.9324089130335453 -0.8859593598754242 -0.8521769289058349 -0.7995738886877699 -0.7073794635864683 -0.5727004448510801 -0.4032810524983431 -0.20868974366963516; 0.1366172766148176 0.24344292503731443 0.3053480492645164 0.32240721625457497 0.2959352023152686 0.22136313848641317 0.09235817683565777 -0.09975387314171677 -0.3682812761404332 -0.7240863416128214 -1.1514627918183478 -1.5878478144847208 -1.953351919181708 -2.19951486470967 -2.314311685002281 -2.3134011222823703 -2.243288725194854 -2.1662988422343195 -2.1256137119792107 -2.1038861871999703 -2.0357563769943345 -1.8729531553445429 -1.605887893667882 -1.25486187940473 -0.8751587318564498 -0.5612623415227634 -0.41167757666859683 -0.4879164376618872 -0.7722440271421237 -1.1841550316883496 -1.6154141402703526 -1.9756285083009815 -2.2059219341698797 -2.283254487398584 -2.2212435035662943 -2.0561308473471773 -1.830361721144688 -1.583704540497364 -1.3544597381726788 -1.174016191483081 -1.0533707591688477 -0.9797866630004516 -0.9251455812974558 -0.8566054475869208 -0.7504400755364776 -0.6021797278560671 -0.4205822265114312 -0.21637251412903613; 0.12030972367199244 0.2068659902580087 0.2425623532856751 0.2285948399952198 0.16977165930668947 0.06687190930615551 -0.07851714340919926 -0.26595946076391574 -0.5036406268403386 -0.8056957093353367 -1.1684385035511993 -1.5507368632118306 -1.8976827979629223 -2.171804528750572 -2.3544306092920793 -2.44760010914879 -2.4743656836435863 -2.4664362553702515 -2.4443833192355338 -2.384050219105584 -2.233642701966172 -1.962764437469049 -1.5676274231921727 -1.0662161689353742 -0.5286194658670803 -0.07926693527449348 0.1472931401618487 0.069610787681588 -0.2750067969353551 -0.7822713821512688 -1.325435880922716 -1.7976773780122421 -2.131943760232113 -2.298395026480482 -2.3062601530434854 -2.1922930785628028 -1.9998417228752352 -1.7666168677387646 -1.5297451155094508 -1.326511408958552 -1.1777459745079382 -1.0774226840813552 -0.9999187521166552 -0.91253091554412 -0.7903968549720788 -0.6279925141304015 -0.43472193336272036 -0.22224027400457821; 0.10096282755632041 0.16482762220640004 0.17204735552238748 0.12344352535799734 0.026180421793775308 -0.11299180729372348 -0.2813524212918047 -0.46450495216696214 -0.6613487638896579 -0.8876007575675883 -1.1551293794366704 -1.4528937990352886 -1.7590658143525797 -2.0522308546935224 -2.309226791097638 -2.5146582313122336 -2.660769212086238 -2.7427365820670455 -2.7534638597610126 -2.6630675446924745 -2.4359314688897786 -2.058622928401023 -1.534091278291424 -0.8845321287394118 -0.19609923837538473 0.381899794403351 0.673192745040747 0.5835487666944573 0.17886150736864392 -0.40627568371890826 -1.039116727748548 -1.6052308855491815 -2.027432675967157 -2.27189632121194 -2.3446368573334087 -2.2811892172993447 -2.125530938816909 -1.9133604817492853 -1.6790122338025357 -1.4627557440563692 -1.2929046309136973 -1.1693854376265769 -1.0698543045637277 -0.9633783719572467 -0.8250305729841014 -0.6489100861161337 -0.44522145278790953 -0.22619674048776672; 0.07971770144641328 0.1221029010780982 0.10539742503532162 0.02729663473821982 -0.10563992463837159 -0.2818165118972023 -0.4768196034262253 -0.6610119263635738 -0.8216658192975521 -0.9716173726976829 -1.1344069041629892 -1.3287335925969792 -1.568800799189683 -1.855528056789085 -2.1672710691092916 -2.471876218339491 -2.7298832150380323 -2.9024377051260095 -2.960388055354736 -2.869145957621846 -2.598601373684703 -2.1469460034707883 -1.5287605887158424 -0.7768040649613642 0.015138609975686126 0.6780417079896411 1.00332799358495 0.899342008868032 0.45796456550514114 -0.16443920574637927 -0.8355576611922593 -1.4493165076433558 -1.9249432400484612 -2.22331252936862 -2.3477417094172925 -2.329565115128555 -2.2094250772466615 -2.020039777462398 -1.7930409849210325 -1.5702109386266718 -1.385716766023969 -1.2444786523386997 -1.1266225281362163 -1.003045376625008 -0.8500928140728525 -0.6624716724258377 -0.45083955067124437 -0.2276171331209787; 0.05693371075358788 0.08120115507261903 0.049866640652925474 -0.045095668683578796 -0.20117379094134427 -0.4049773071547929 -0.6239930291272769 -0.8168383869032813 -0.958307176721995 -1.05244921543149 -1.1256796755893042 -1.2169233044236838 -1.3713014218640223 -1.6173826256400283 -1.9442202317849138 -2.3071391987845167 -2.640363626918016 -2.8829131013391716 -2.9918885139156024 -2.925213932559811 -2.653797459272418 -2.18464732794204 -1.5480889820083539 -0.79174504564197 -0.009570682138008966 0.6356604585518585 0.9513573652314217 0.8526609970390425 0.43685466715763405 -0.1469263329407653 -0.7802426763521922 -1.3708790022187924 -1.8449230282775766 -2.162385639246807 -2.3197384974607105 -2.3385796983189 -2.251176400216969 -2.0843444535257656 -1.8666475553894097 -1.641160849031484 -1.446871689704236 -1.2932062790011947 -1.1624951203094689 -1.0264159436107774 -0.8624826423585655 -0.6667697228524245 -0.45069289809067964 -0.2264694539464854; 0.032352254459325804 0.04139267422742126 0.005246549893379406 -0.09074313359347397 -0.2514056866099183 -0.4664467748070296 -0.7017420293582247 -0.9094144168087984 -1.052166132625231 -1.1205159880580828 -1.134407130324271 -1.1398571117573433 -1.202598253198578 -1.3782625878123829 -1.6734402796187733 -2.040651420689125 -2.399222992868402 -2.675180084704038 -2.8159740184967195 -2.7765216974654447 -2.53213884364889 -2.0998195766751175 -1.5360317619003054 -0.9006238919604664 -0.27494814494630226 0.22345351081290651 0.46573585967099396 0.3990347181614421 0.0799510074205744 -0.38081832380770875 -0.8922846150252326 -1.383078054644027 -1.7927012365715131 -2.086084420866669 -2.25287894245496 -2.299209086497441 -2.242328679100771 -2.099496728152742 -1.8947770872089411 -1.6715158751935153 -1.4725781740153248 -1.3119204622844922 -1.1739505510955575 -1.0306751116743573 -0.8607156328259828 -0.6614427501509437 -0.44479948826236326 -0.22269544413717313; 0.004942218684542141 -0.0011190327528098557 -0.03625524306790998 -0.12059451740095681 -0.26803311943212116 -0.4766867513524427 -0.717938831163769 -0.9421668093978288 -1.1017323056579493 -1.171968886261115 -1.1601489498630875 -1.1085948982471514 -1.0904648756314614 -1.1799564312035744 -1.4043066625412808 -1.7242706520577835 -2.0556139980528094 -2.316203086716108 -2.447353317152158 -2.406321963538988 -2.178527993831151 -1.8126072617868219 -1.396581717615424 -0.9800377634025571 -0.6088266107164986 -0.3385348870857828 -0.2256348433992998 -0.2903197837697481 -0.4988067447861335 -0.7946394470622564 -1.1298946564870698 -1.462119174017231 -1.7556187462106627 -1.9877388163608558 -2.1409302468916462 -2.203607293762411 -2.1746964029562914 -2.058165462698516 -1.8719762011446934 -1.6585491557618162 -1.4625061556072938 -1.3010715074220633 -1.1611698251743827 -1.0158340167458155 -0.8446451392229118 -0.6463242347022874 -0.43312695771559145 -0.21630581470167864; -0.025750161954848264 -0.05045605183229241 -0.0862080103583004 -0.1554302680811847 -0.2800852197924094 -0.46920529742022277 -0.7043193246181239 -0.9388663764721058 -1.1193104134370355 -1.2084648094906203 -1.199328740548699 -1.1235726106974133 -1.0499209326297516 -1.0588049846696597 -1.1943099708838865 -1.431173549725592 -1.6893172656279316 -1.8834544822926529 -1.9529629379669102 -1.85900890770618 -1.607813163627223 -1.3019811123444573 -1.062659739090249 -0.9032533268796333 -0.8134178964675215 -0.7930917871390663 -0.8433125492994816 -0.953750844279336 -1.0941935619708165 -1.2446972710286215 -1.397255851262067 -1.5492231755256676 -1.7015079218251232 -1.8505959870640152 -1.9757524512530615 -2.048164359766686 -2.046333306745789 -1.9591622521185121 -1.7981788168035602 -1.6034319391318441 -1.4189085712247167 -1.2636808789768872 -1.1271956151215028 -0.9843425857563204 -0.8164743633537972 -0.6232925706149676 -0.41678269188739897 -0.20778460454888267; -0.058557721224185955 -0.10751539858135774 -0.15286410511981618 -0.21526489586415057 -0.3201305474728689 -0.48502153680222654 -0.7026852353754427 -0.9342978521166115 -1.1262465293014257 -1.2345235151838196 -1.2419265810564208 -1.1682054561435609 -1.0704292610105501 -1.0230900897289625 -1.0760788427706744 -1.2162471735158071 -1.373341326616904 -1.465892601819928 -1.4352494355675958 -1.2483992644035493 -0.9372728362270042 -0.6593423465953179 -0.5733943544682872 -0.651307381367185 -0.8167774577688307 -1.022451073870378 -1.2368329540395289 -1.4263694772783857 -1.5545726340787704 -1.6083476637852077 -1.6054801755876458 -1.5882416158459445 -1.6012359189718555 -1.6633441422923523 -1.7540675387090565 -1.8323663000073755 -1.8581678063245826 -1.8048027705055925 -1.676867123169115 -1.5108336096798984 -1.3474846802121256 -1.2057120922920022 -1.0778375735843444 -0.9417789755524006 -0.7810690969828419 -0.596068989000593 -0.39831836098408574 -0.1984292950231127; -0.09181595198000092 -0.17062946959072284 -0.23826053188513657 -0.3105833506176558 -0.40995441595537346 -0.5562646483019815 -0.7511548360734519 -0.9661152553122968 -1.1523997043047611 -1.2650429088533615 -1.2834314230904713 -1.2206269429888015 -1.1218078045176805 -1.0475761670869563 -1.0409187235243442 -1.094091427746149 -1.1488791443562387 -1.1345123628941027 -1.0021578796856643 -0.7280961434523194 -0.35829217706798233 -0.0795693937314349 -0.08619002223616806 -0.3295955714245607 -0.6761727111538451 -1.0440891006043616 -1.38810967778557 -1.658662050637323 -1.8065437228951435 -1.8136313854501738 -1.7095945434682986 -1.5676325738941892 -1.4664304434294928 -1.4463231654187951 -1.4957086502836603 -1.5709119927025754 -1.6189904215307598 -1.6009340061536923 -1.5139341799955122 -1.3871118807400926 -1.2544816638533247 -1.1336110529696923 -1.0197495975509134 -0.8944112352470213 -0.7437492935297307 -0.5688331558339222 -0.3806267811127445 -0.18963449323761458; -0.12518826571102318 -0.23811143337996563 -0.3391392139228528 -0.4389409130981646 -0.5521579471981963 -0.6942760679544783 -0.871123516282695 -1.0638226359522567 -1.2304929079778693 -1.328589340180389 -1.340174430331979 -1.2798820662852695 -1.1853332190756514 -1.100889901813621 -1.0558287705625222 -1.042507852700673 -1.0155055221559819 -0.9188608526370008 -0.7175806866937761 -0.4015869073328029 -0.017444941495410524 0.2587561642181957 0.23159461489911537 -0.0692427084984105 -0.4859046241608777 -0.9243344683868929 -1.3335082680826993 -1.6551000711005759 -1.8303261847463996 -1.8358721835325598 -1.6996129993254654 -1.498613061122036 -1.3234078303333738 -1.2333408632353384 -1.2345154013142905 -1.2912274783295683 -1.3484431090476203 -1.3618988160345926 -1.320420490660362 -1.2406953720567637 -1.1466652207954133 -1.0531977827608519 -0.9582032380994091 -0.8471786415320943 -0.7089095371851023 -0.5450884962119831 -0.3662309403499316 -0.18289355851519387; -0.1593036864577879 -0.30892144098540314 -0.44942721793099427 -0.5875311554172699 -0.7295889729829321 -0.883222497135726 -1.0531503356768517 -1.2265906466525907 -1.3684720791483092 -1.4404712614771318 -1.4300770497918271 -1.3574833195778382 -1.2575235896987464 -1.1621001185418527 -1.0880611780877363 -1.0262000280585815 -0.942043573055566 -0.7967208704019563 -0.5731871581568799 -0.2752640290480686 0.06210122776890201 0.3105188409451561 0.31069195424163903 0.059065891098801296 -0.31249122400511387 -0.722950784224275 -1.124574112372712 -1.4582170180093763 -1.6612290087393562 -1.699860026886665 -1.5889109557017747 -1.3884045242429504 -1.182121263405509 -1.0399954588125198 -0.9894221988750943 -1.0141767439041873 -1.0684991037372829 -1.1082780605758271 -1.112909814172093 -1.0833193044295257 -1.0315814185830945 -0.9696484878250858 -0.8976053280757946 -0.8038922781240557 -0.6797846415896346 -0.5276784773605627 -0.3574089520269945 -0.17949501641523516; -0.19317583007642014 -0.38010506604257394 -0.5614453388810285 -0.7402966921295918 -0.9163773033430367 -1.0897974438432056 -1.26118703882225 -1.4198167481329125 -1.5367161921478811 -1.5797452309339655 -1.5421677917825676 -1.4481765277485845 -1.331303801691324 -1.215686879919892 -1.1106705812695545 -1.0087187111378677 -0.8865656522163144 -0.7190246242600147 -0.5047475683096849 -0.26374801102100304 -0.01816513372867049 0.17017791232372015 0.20469571145411306 0.06358982267849955 -0.18372862103630352 -0.48866840228791697 -0.8210432283369375 -1.135027719844024 -1.3646236095860491 -1.4558823639180476 -1.4016447323187595 -1.2377345529776254 -1.031744697539152 -0.8556162121946799 -0.7585225860480829 -0.7487734063477328 -0.7962569893958011 -0.8600670724174796 -0.9083769904089296 -0.9261792605899825 -0.9160132156494206 -0.8871489783196783 -0.8403086660083745 -0.7661085998869839 -0.657773820496731 -0.5178168388225968 -0.3551355158391847 -0.17996713341362747; -0.2237484612817865 -0.4456288039426805 -0.6656380693508261 -0.8822034755813333 -1.0895771563661893 -1.2823691936955748 -1.4568803669820403 -1.6022250458024072 -1.6953488980616584 -1.7118330288197532 -1.6489630435204876 -1.531518015668307 -1.3914192672837635 -1.2485165611375675 -1.1078500901006625 -0.9648239238164907 -0.8102299810385748 -0.6347796747263789 -0.4459250457179201 -0.2725506501963703 -0.1307629817090205 -0.026031305519230972 0.01558182368829506 -0.017678699144180146 -0.11146055531314752 -0.26476900172999385 -0.48875111181025704 -0.7622160672294829 -1.0090750673105633 -1.1493152767181076 -1.1525302002116 -1.0340091342134514 -0.8436272678188074 -0.6523237189804769 -0.526791050072773 -0.4957069624617655 -0.5439917898065935 -0.6322231799167998 -0.7179685823426512 -0.7759036825039025 -0.803112400874596 -0.806338607469417 -0.7856920607882115 -0.732889001176604 -0.6419989071126905 -0.5148266088073593 -0.3589127522945865 -0.18404061366424676; -0.24900355726310577 -0.5001050537190213 -0.7527927501384571 -1.0008224556922005 -1.2332623575450476 -1.4405807872290883 -1.6161063061385603 -1.7490496384691554 -1.8216139129379834 -1.8167157903533526 -1.7341581465693792 -1.596631134179598 -1.4330195045038874 -1.2599469690684357 -1.0802220317827058 -0.8927083774654322 -0.7010456369645984 -0.5142889982609424 -0.3501142636211385 -0.23642110047444828 -0.18328495053723112 -0.16457688432197715 -0.15069686500448523 -0.12706529213682274 -0.10132199730991169 -0.11519816768734127 -0.22653243612710167 -0.4394088471050424 -0.6713082309864233 -0.8257716500327009 -0.8571052190292898 -0.768057115724851 -0.5965221908069226 -0.4115336310853851 -0.28617112776450737 -0.25832994197173603 -0.32001120772891256 -0.43072083642154463 -0.5443414399817683 -0.6333395263929554 -0.6925769678206964 -0.7259909020718951 -0.7319446067692834 -0.7024041452270473 -0.6310408648212533 -0.5176066420542311 -0.3677115567991183 -0.1910353655184719; -0.26945728992717427 -0.542775753143367 -0.8192975552892933 -1.0900133343462666 -1.340007536712354 -1.55645617186353 -1.730960357954687 -1.8540794796640845 -1.9122265044276214 -1.8929142217311425 -1.7969999727623316 -1.6445873731973468 -1.4616082384262423 -1.2618669201252906 -1.0454429862875998 -0.8133297761011967 -0.5798999313043762 -0.36903624314343575 -0.208505636238988 -0.12483559212612591 -0.12104255547980369 -0.15991663887396435 -0.18704703256847788 -0.16862923344688052 -0.11091096926970641 -0.06492949203631458 -0.09944054548813545 -0.23221675565967154 -0.3991551792873078 -0.5156782119891281 -0.5358605848749362 -0.45394394245585773 -0.30108906516437794 -0.14259784082776544 -0.04552988086441057 -0.042596468512072605 -0.12400940769228101 -0.2516312239440153 -0.38435387835932 -0.4975022896178668 -0.5847736096140417 -0.646592924502461 -0.6794457715927963 -0.6749148387512043 -0.624630148030455 -0.5252067685099652 -0.38033713670942065 -0.20007031978707407; -0.2869783571782132 -0.577055254277088 -0.8691556079860077 -1.1534580785214184 -1.4132586262914921 -1.6336980404641968 -1.8057810351168813 -1.9220925870964405 -1.972519096685644 -1.9465642417647295 -1.8436489334845307 -1.680265092701539 -1.4804258983917908 -1.25843792007401 -1.0147573019174063 -0.7491420969992595 -0.47912943405098757 -0.23728769263972146 -0.05595326717805752 0.03892225572587275 0.04511263290961239 -0.002871090807533609 -0.05141287409155152 -0.06182400347936254 -0.032033839051749384 0.003171537757874315 -0.0037306999974490948 -0.06974471577834078 -0.15738493752114244 -0.2125640338971603 -0.20070476881384752 -0.11536612247245297 0.013950716040771808 0.130724402098716 0.18386430202756493 0.15361494169556528 0.05224327609527117 -0.0886542652419793 -0.2362567236470763 -0.369987846724544 -0.48251592263492843 -0.5712798199008197 -0.6309536974070916 -0.6517135527771266 -0.6221773015563091 -0.5356986236460048 -0.3945276097723777 -0.2096267638432922; -0.3019514736027138 -0.6054218274849714 -0.9082929953906415 -1.2002702516714763 -1.4641745156445303 -1.6842680383491555 -1.8518459134105196 -1.96200100332522 -2.0081218021113494 -1.9811581281358066 -1.8775549469505215 -1.7080583511984497 -1.493714573599621 -1.251815919371448 -0.9886781371149413 -0.7052675794541915 -0.41438901984255405 -0.14720100388441942 0.06444326650248287 0.1973360731980896 0.24476370953519314 0.22867598996287405 0.18940749382588332 0.16162246523971105 0.156041731583224 0.16007628014104858 0.15314300221791827 0.1270220267673759 0.09961371577763371 0.09684330472590019 0.1387457151391514 0.22437440625436944 0.32475037975551124 0.3952621176848671 0.4023545250602149 0.33802162690395393 0.21586599502666612 0.0610664635123387 -0.10137268213139573 -0.25460330684802246 -0.39061715521300727 -0.5045589820182033 -0.5892724207886412 -0.6329770406990161 -0.6214061161791008 -0.5455668493021208 -0.4069725853514754 -0.21778431062698053; -0.3130072651400551 -0.6267408282289029 -0.9376895097681367 -1.2347213097475758 -1.5005407936608135 -1.7188563057680217 -1.880682711582918 -1.9832876150738892 -2.0245162111076986 -1.998572984378421 -1.8992780800148297 -1.7299768694125008 -1.5065820612299794 -1.2486745034278066 -0.9705706805017714 -0.6796305977166063 -0.38424252780583856 -0.1059350433922013 0.12855828372132888 0.2999968938496521 0.39911660849379177 0.4338483434642827 0.42743705470048954 0.4066827187537844 0.38839533247121016 0.37731453607215165 0.37060053628538964 0.36820083189338704 0.37755628407590813 0.4098055948423441 0.47135705956918933 0.5511985541348513 0.6208092257532425 0.6473554808943863 0.6117947500370142 0.5144752345846069 0.36981315633764017 0.19796955903850993 0.018244988483747136 -0.1550702595996567 -0.31320469434672044 -0.4495861871495097 -0.5557036110980006 -0.6177365406279185 -0.6194074115247188 -0.551063345173219 -0.41437540578939086 -0.2226756700549366; -0.31984599722392754 -0.6395953173787701 -0.9545991449566179 -1.2536285327862382 -1.5203134977952941 -1.7382075500098766 -1.8964253252078045 -1.991950973095053 -2.026394352206188 -1.9998031829338438 -1.9069248235987164 -1.7440315418890062 -1.519549498016933 -1.2527708705146834 -0.964723385375725 -0.6714448176847684 -0.3821719234074889 -0.10846855432303985 0.13234337722183734 0.32614085842287616 0.46446887191689845 0.547476782653946 0.5852831191935713 0.5952348108109673 0.5948815734353223 0.5966135523371808 0.6059821202575687 0.6269099212231025 0.6631588217807686 0.7187173266810363 0.7888734057837768 0.8561676566742153 0.8946968852441133 0.8820930677217381 0.8094351231907332 0.6820623531483493 0.5142764102026438 0.322776277911773 0.12323411333614258 -0.07112225532214156 -0.25023911173602037 -0.40610896184638995 -0.5287979079134097 -0.6031080538650877 -0.6127593893193934 -0.5489356517829017 -0.4141773910378887 -0.22284788990066198; -0.3231430591929813 -0.6441447331968301 -0.9574587264720205 -1.2530683704373027 -1.517522427893029 -1.7356513744830184 -1.8941512014623418 -1.9866401145993953 -2.0150331027914614 -1.984590863376149 -1.8955405903301707 -1.7424099118483187 -1.5261357923665333 -1.2618608279145032 -0.973224445680699 -0.6830328662179077 -0.40546578021918905 -0.14746652283056833 0.08346766760740863 0.2803476528749685 0.4382811230900982 0.5554935869940929 0.635861812828952 0.689117267622566 0.7290012751766484 0.7672672454952909 0.8111777992592305 0.8643190792224398 0.9284669052607191 1.0017327485155338 1.0735779779096999 1.1255966371803645 1.1367052095499943 1.0926039089724788 0.9906099253678488 0.8382881972999425 0.6489516968765172 0.43677357977465114 0.2158367241469959 0.0002331454983911925 -0.19806847165715727 -0.36949793054832375 -0.5031364566224952 -0.584184369637198 -0.5979000149339647 -0.5368754246808088 -0.4050189514404575 -0.21771596491707831; -0.3221123494654594 -0.6394559524288023 -0.9456561514059857 -1.2325925770713795 -1.4905906326610268 -1.7068772756585753 -1.8669525373760179 -1.9606290987401687 -1.986941605395619 -1.9524520695363898 -1.8632972928322329 -1.7189378313156298 -1.5180876547002387 -1.269875589853933 -0.9947792264084684 -0.7170920070632315 -0.4555320503165082 -0.2179795951343093 -0.006247606166028772 0.17956807468566008 0.3399177353407664 0.47482163479728573 0.5870237132617349 0.6827671340760464 0.7707681082439802 0.8580403104301526 0.948502831651037 1.0424956402857901 1.1381566984800306 1.2289262074126823 1.302910194085181 1.3438552437420197 1.3360514207123677 1.2709059886801686 1.1490220303167271 0.9782797996452186 0.7704204061385623 0.5383890309806686 0.2971466962833009 0.0627515934851468 -0.1511532896238593 -0.333461443605513 -0.47286859851191104 -0.5564552488167772 -0.5718580573510066 -0.5133969419562358 -0.3866039834566813 -0.2075009242923673; -0.3138569784651194 -0.6219403276964259 -0.9173241373967674 -1.1928893646472445 -1.441413976551365 -1.6525709938502218 -1.8122110818562185 -1.9082650405952912 -1.9367228920557715 -1.90233154097478 -1.8136972587482456 -1.6768445645412102 -1.494403957506505 -1.2724681744760078 -1.0253056791791781 -0.7730641106342767 -0.5345052471192618 -0.3196724042039266 -0.12919286778928013 0.04078201351991183 0.19472427725486038 0.33569230269890865 0.46764259235024025 0.5964874483227325 0.7276491424324031 0.8633383952275803 1.0019396581397637 1.1388467241761084 1.2673979796093722 1.3780322027396597 1.458897093232588 1.4968448977120927 1.4810499274673312 1.4065374074944899 1.2750440330691015 1.0934909453882855 0.8721491495453673 0.6243443192068747 0.3670714435274229 0.11841109014382818 -0.1062491524896067 -0.2940607225447612 -0.4341078073922332 -0.5165951784930237 -0.5322680110446092 -0.4775546075752635 -0.35916379761915096 -0.1926408458913208; -0.2961912618487893 -0.5882741295971391 -0.8697493907276934 -1.133044957090356 -1.370681830528381 -1.5736347389443437 -1.7297963170373583 -1.8276572869427832 -1.8613720792399462 -1.8329184630827315 -1.7503552429243878 -1.623546460693742 -1.4607082490403667 -1.2692754520626492 -1.0592737567596353 -0.8445080579283188 -0.6391543468215021 -0.45193258853473633 -0.2833310515855127 -0.1282179746210433 0.019416008666643666 0.1638185470412226 0.30915324555456153 0.46133098820373497 0.624548149267478 0.7980653331166785 0.9760558013948782 1.1495871876069608 1.3079309920744289 1.4394660255299914 1.5325504754610044 1.576664841163925 1.5639695307699237 1.4911534124529515 1.3597031284425158 1.17496391991308 0.9465810525066123 0.6893499222353828 0.42259436447960347 0.16630911991965475 -0.06228348605205185 -0.24912299999951498 -0.3847665778429082 -0.46313068429912313 -0.4786816712683156 -0.42984800188061295 -0.3232660784975869 -0.17326336214966273; -0.2699811869276096 -0.5393898705342277 -0.8030910028329904 -1.0522842828017613 -1.2776103871149183 -1.4699868237578884 -1.6201108865103984 -1.7190094652235202 -1.7603753643535078 -1.7432764356784378 -1.6737700474878876 -1.5626157233107325 -1.4214465081339729 -1.2608276209670495 -1.0902420445385665 -0.9189166037007073 -0.754995335307948 -0.6023110033393263 -0.45907491254256494 -0.3195344456023451 -0.17744696230529763 -0.029237367520193916 0.1275873776209184 0.2970951086751379 0.4823025942714103 0.681084325919085 0.8859036719275221 1.085780056846331 1.2681126200322486 1.4200608367009961 1.529760018755135 1.5874102820958733 1.5859099309827798 1.5220921961294476 1.3965914736590206 1.2141836559532622 0.9853906274451861 0.7267561574670541 0.4592583947428017 0.20420429351262495 -0.019868811760059807 -0.198932922213684 -0.3258358000937118 -0.39816389635023486 -0.4138247457453339 -0.37244667051787866 -0.27999663469009745 -0.14978032447571232; -0.23905023148682558 -0.4809844399930067 -0.7224318947777467 -0.9538647340835693 -1.164189980464256 -1.3439892708485408 -1.486095350996512 -1.5844288226527463 -1.6337271898779713 -1.6312744798360002 -1.580573190719751 -1.491268456525578 -1.3754824610558132 -1.2455973606891617 -1.1123658138203512 -0.9832905657706938 -0.8618671632422721 -0.7466277955443835 -0.6318813694432917 -0.5106414968496662 -0.37696806375730024 -0.22840417225635193 -0.06454755064857491 0.11605795098844411 0.31472573001509024 0.5285136332531772 0.7497062185395879 0.9672775828079103 1.168254574236889 1.3388491054159541 1.4662594707831222 1.539738615264872 1.551576157227933 1.4983322638263257 1.380873926415602 1.2050662188915398 0.9828182601685128 0.7317659492924513 0.4732535398334324 0.2291487046801644 0.01838328473471521 -0.14656573635626666 -0.2613101629753686 -0.32622082248165757 -0.34147130090540595 -0.30770954481048196 -0.23071496312912096 -0.12290925562278224; -0.20768133225876328 -0.4199701834835653 -0.6349929753101458 -0.8441354531658237 -1.0360211985634884 -1.2011448523244124 -1.3332521161117388 -1.4281073787332141 -1.4822511736541513 -1.4935377759284372 -1.4639809548541658 -1.4009401614081614 -1.3147002723423333 -1.217007258219762 -1.1188623984787835 -1.0273032053878821 -0.9433263027363703 -0.8617988477235428 -0.773823036140135 -0.6712799010826496 -0.548837107448498 -0.4052123797790289 -0.2417349206364301 -0.05872948383047325 0.1439526204976251 0.3631692240393872 0.5916660026687811 0.8190083863517231 1.0320002367622856 1.2159213529623287 1.3567046268473715 1.4423499504512196 1.4646101063003751 1.420454896011227 1.3121353697662486 1.1469797295303221 0.9379790281540454 0.703066453520475 0.4628943358876979 0.23901644584608 0.04938522530876077 -0.09631149814675441 -0.19615597075508534 -0.25179271728898645 -0.26489516172834743 -0.23801012096954882 -0.17740095281784132 -0.09394965176825294; -0.17774501224846465 -0.35989106906952434 -0.5455538834689729 -0.7283131420511912 -0.8982870070373679 -1.0462066044855836 -1.1659249903988507 -1.2537806667798033 -1.307957392101286 -1.3284870945752016 -1.317928484887601 -1.281594978881776 -1.2266475129947216 -1.1622664670139056 -1.097790343448969 -1.0389353638332846 -0.9854607386918237 -0.9306062717925808 -0.864242173722897 -0.7781474574552075 -0.6679485534904258 -0.5334836767386777 -0.37721518633251677 -0.20034849589581147 -0.003144965540188124 0.21163355591405675 0.4374681446808427 0.6644319092479246 0.879146410532755 1.0664145600436659 1.2115561384124454 1.3021812360145222 1.3300324395469796 1.293022336394828 1.194893664694955 1.0440453749391672 0.8542417539191833 0.6430739126053728 0.42974763266923494 0.2339894699977737 0.07111899001822396 -0.05194699617856187 -0.13473322315619998 -0.1794434318720541 -0.18925392775417696 -0.1686533968237703 -0.12437193667612391 -0.06529971482419668; -0.14826841304085772 -0.2999304805371118 -0.4546805733429123 -0.608160217327476 -0.7528425912134465 -0.8805817417933729 -0.9854136318762404 -1.0633256342835256 -1.1133222525959612 -1.1378799312244572 -1.141298858334812 -1.127959232124893 -1.1020436889590124 -1.06916289335574 -1.0355737129852562 -1.0046940602208203 -0.9751416258896626 -0.9399818104540373 -0.8898286539788104 -0.8178285531090325 -0.7208152838307677 -0.5992904312211215 -0.4560764724707375 -0.29268514602645557 -0.10928882159274599 0.09161080519756083 0.3040543752010966 0.5185622749034037 0.7222023170390265 0.9003500977860476 1.039115762919931 1.1266595977154394 1.1551373374993548 1.123464104404958 1.0365791762810705 0.903666702439482 0.738668182905553 0.558062548144934 0.3785443759128101 0.2163012485578831 0.08327740794350928 -0.015659821807532132 -0.08084094337725567 -0.11487948693793336 -0.12190180337442884 -0.10736763906801347 -0.07786585404871993 -0.04031479762194776; -0.11793519322403423 -0.2384328803989873 -0.3613552289670682 -0.48381886284781417 -0.600450509553389 -0.7051139852113426 -0.7927093487687711 -0.8587680604522868 -0.9019498886527617 -0.926315629496913 -0.938074753727387 -0.9414228431242774 -0.9384156393306089 -0.9314006940602265 -0.9231431199194083 -0.9144859462024086 -0.9028434240594994 -0.881876522527911 -0.8440208790574578 -0.7847017006290006 -0.7023102728640048 -0.5976915597082458 -0.47324421075785006 -0.33020107615828165 -0.16867135530055727 0.008863059453249639 0.1968219024900634 0.3863552631664639 0.5658323968236626 0.7226045414141719 0.8449032097941492 0.9221905260536993 0.9474797436614913 0.9205868404052103 0.8472898120848896 0.7369512010113191 0.6022687521005864 0.4574040569713124 0.3156580030482871 0.18903951388416865 0.08637155088642873 0.011163525321396469 -0.03737843935292618 -0.06224512448030075 -0.06777767859357176 -0.05915832473922316 -0.04193795866264828 -0.021216092870732776; -0.08695505394452947 -0.1757993877115396 -0.2665255106531525 -0.3572691028269401 -0.4442311716850494 -0.5235896169611233 -0.5916298564365791 -0.6439731917982723 -0.6788488502974519 -0.7004108505670092 -0.7153389952349422 -0.72771815773915 -0.7384306272163551 -0.747743253515815 -0.7558987088055569 -0.7618877453046798 -0.762408130949495 -0.7518668927469466 -0.7246337508583911 -0.6783157577228534 -0.6127675686487736 -0.5290107912152245 -0.42885116393951545 -0.3131006861630086 -0.18196450306956669 -0.03770461331551645 0.11467978512586598 0.2675570324992947 0.41156723206463935 0.5370870521123153 0.6351702156408768 0.6972980402899832 0.717685091277312 0.6967209103525256 0.6402672443259979 0.5566502248586894 0.4559204313096065 0.3488499142999504 0.24519731231077313 0.15346170526837366 0.0798932266314206 0.02685937470763746 -0.006680544175583249 -0.02395687769408332 -0.028927124234408193 -0.025408908186634462 -0.01733135744816064 -0.008319386481941752; -0.056558397333751266 -0.11424516918141693 -0.17340592152564718 -0.232882573372475 -0.29003000831405484 -0.3427440662021621 -0.3889755950294292 -0.42532734430285585 -0.4502904524173348 -0.46705363763768376 -0.48077442878882537 -0.4945481756968654 -0.5087401116009895 -0.5229407144271689 -0.5364925539725836 -0.5478647188190304 -0.5540467126536719 -0.5505649153814908 -0.533524541204103 -0.5018171566114688 -0.4560522203587118 -0.39723933958271096 -0.3266842961705442 -0.24498668685660543 -0.15241876568497875 -0.05070459690506931 0.056430634373944466 0.16346015136556438 0.2639145527926998 0.3514982472123411 0.4202896644467389 0.46419678403894316 0.47887716284821 0.46489625658759615 0.42713303634047234 0.37173152810367177 0.30548940674186914 0.2355137650623722 0.16826702375488123 0.10933240871036093 0.06264083875298937 0.029482409891146533 0.008713094175260332 -0.002426731094645517 -0.006747140409056985 -0.0063680482774628 -0.003699109265987947 -0.0012687124337201143; -0.027634405541235373 -0.05579161698188116 -0.08479320535411382 -0.11405572882772506 -0.14226025360330363 -0.16829951696678142 -0.1914210859138068 -0.20995925860311 -0.22311276355776402 -0.23255305672023133 -0.24095028821490339 -0.24978045270068902 -0.2591304049050145 -0.2687221953892017 -0.2780819721100764 -0.2862496047815587 -0.2914724750551905 -0.2911921234160476 -0.2833241843084404 -0.2673793954233863 -0.24391207749368984 -0.2135532148875684 -0.1770335918147478 -0.13478037325365222 -0.08696775875021083 -0.03449280490365631 0.02071800116566999 0.07579360619442303 0.1274673862681757 0.17266629140807047 0.208388868356047 0.2313582218075804 0.23921239283014786 0.23229912356065577 0.21340359250202082 0.18587127765851022 0.15312303444478992 0.11864793209016192 0.08575950377321802 0.05722625391040994 0.03480566571323327 0.01899036719449954 0.008999181395346416 0.0033208878061386227 0.0006593671281814491 0.0001776179744554502 0.0006926937179713032 0.0007960894439121046], hyperparams = 95.59306645075016), (params = [0.04734979794646056 0.09085718464864447 0.12496045449154738 0.14300907303426835 0.14657831594378387 0.13947252325775727 0.12396408566722408 0.0985852707526157 0.0668688771643293 0.033699998743666915 0.005985087722671267 -0.012717669026532173 -0.023920673396111418 -0.025721617728406084 -0.016239562219197513 0.0010114137179957437 0.028308980699517594 0.0601402241966695 0.08668840948553495 0.11068739514107295 0.13275309424779902 0.15171570131701145 0.16743167131338557 0.17858243598148782 0.18111603647984262 0.1729657378534728 0.15666489260643293 0.1330784730435229 0.10137106473957637 0.06528861793152492 0.03197082471588381 0.004464116372067288 -0.017518842737226984 -0.03382673433122877 -0.041039712362719055 -0.0337762441172042 -0.01147677086065401 0.020811790601629555 0.05376008621723068 0.08051452891448664 0.09968480891310177 0.10909582584874827 0.10488615684399474 0.09092768434760687 0.07176346364454507 0.04958815637781112 0.030285864337867684 0.014945429092312144; 0.08400723853386556 0.16405506131602715 0.23125181385559174 0.2726934011445376 0.28410211235732613 0.2736155362973164 0.2462293048850238 0.1993508076380499 0.13878561564525865 0.07508825836178627 0.023436905120085765 -0.007985418764628584 -0.023929666234256672 -0.023851539136207148 -0.004945567611109992 0.02941054268665454 0.07982739370285576 0.14018386385307563 0.1945541434235551 0.24516125798460348 0.29048393116438387 0.3294766279588963 0.3638967395337129 0.3858165672703741 0.387994050217292 0.36639438792424056 0.3252021226695726 0.26973812759481897 0.202941808119985 0.1321620633330085 0.0677741807681973 0.01477511648304728 -0.0275369917063503 -0.0586665557728639 -0.07237178916965846 -0.060050456767751545 -0.020237496903397715 0.03753009303367894 0.0974858799290242 0.14728952348956206 0.18240857081309517 0.19935735244675426 0.1935410878649296 0.17280800198328916 0.141944750152663 0.10236881941794056 0.06422432562025349 0.03135480572008818; 0.11258378604048287 0.22413622849871165 0.3209654121642398 0.3859359212186874 0.40949811529710173 0.40005007917514734 0.3659979247612455 0.30674724539061243 0.22738590531067168 0.14329526003010154 0.07661928694276146 0.038098990139981456 0.019660420036498743 0.020086694128216398 0.045245774127722314 0.09349780510209015 0.16302759830266322 0.2483846741928609 0.33131479435651695 0.40983100084624113 0.48113424806977634 0.5433802290429551 0.5962154457571484 0.6252141685158843 0.6203080225717813 0.5776502038775692 0.5039029760316337 0.410268389945964 0.3050833614617257 0.198864340160087 0.10440886300168593 0.027975445598795994 -0.03183634470252937 -0.07302595558256023 -0.0892140912504788 -0.07348075228264637 -0.026304127981107376 0.04278802834863486 0.11741928825886624 0.18239855468144414 0.22783053307698165 0.24945577889702558 0.2452331249121634 0.22407196327301587 0.19118304980647371 0.14556954069479136 0.09504100703990068 0.04685328378230124; 0.13744163924131425 0.27605749698825405 0.39921755073600423 0.48558640604526 0.5245183869576476 0.5239978907525051 0.49309501688996343 0.43324999219676397 0.3502508191883812 0.25995025674455496 0.18380691744481237 0.1353718013067039 0.1083267170636716 0.1023905541671558 0.12664228757011453 0.1822799231475269 0.2673958187083631 0.37629020583980477 0.49066242630348256 0.6013487269507924 0.7004246804792283 0.7843044937191996 0.8475062525502227 0.8741642691551188 0.8546654514655714 0.7881398649529447 0.6798513148141848 0.5453286331532994 0.3998550820307678 0.25697330670097823 0.13178650989593044 0.032397834132197316 -0.042740777760290026 -0.09137481260284627 -0.10926153010307417 -0.09245685330282827 -0.0451317959161097 0.02481658735892021 0.10346070662855845 0.17507899195763016 0.2257500434624118 0.24903404404075336 0.24774344498201148 0.22924531267427842 0.20152026783656823 0.16120233464376246 0.10936059747148064 0.054716554589370664; 0.16127581506832347 0.3228549252450851 0.46687527401141304 0.5714395301959277 0.6276609925450747 0.6437032614819366 0.6248937254946402 0.5732538014168935 0.49845732597477593 0.4137624225830126 0.3339601700885847 0.27211618771500884 0.22905383183052982 0.21081143605725655 0.22916672791505546 0.2876991005254485 0.38612190555466847 0.5189200772944628 0.6669253998083869 0.8106933767116864 0.9334994618549791 1.0289784637804713 1.0917171596839994 1.10809159086088 1.071313773382841 0.9808675966157645 0.8402408840789529 0.6668914451622946 0.4798854003938076 0.29555910023706006 0.13335280461942206 0.004764625763097604 -0.08792594520975522 -0.14314565668604998 -0.16026306873206142 -0.14196546841045324 -0.09569884193594719 -0.02819454360113651 0.050511440608947265 0.1251548565397949 0.1788385321769669 0.20227804129295074 0.202987230963773 0.1899842682144223 0.1715783828366507 0.14189290770780488 0.09690049710375356 0.04722013965114326; 0.1801811379805804 0.35918711721921925 0.5180380557538221 0.6391635939146041 0.7142739124822045 0.7483603884711777 0.7439976581700782 0.7040342500201144 0.6401039906570598 0.5663950210849067 0.48903039103606133 0.41734467859124397 0.3599863170292868 0.33226724678808217 0.34726220771592486 0.4115212763573947 0.5235704540197914 0.6777910415861795 0.853587091691582 1.0220840465720682 1.158909584651125 1.2563355880827485 1.3131362901723453 1.319720655959584 1.2672056950474564 1.1514702779605632 0.978747982147306 0.7667170504795175 0.5349187849402072 0.3048770820977645 0.10155728253200673 -0.06032068271599778 -0.1739672901371283 -0.23981380304885155 -0.25955699255553594 -0.24129918988297896 -0.19468133203259685 -0.1276333887707587 -0.04610138622776918 0.0358399937706483 0.09647389885230682 0.12265984671746537 0.12759185539117288 0.1260053593779553 0.12051876434497764 0.10056575413920206 0.06457836403575433 0.027832522460065064; 0.1921032890450011 0.3819069740190841 0.5538107912707402 0.6935275611991562 0.787171360772709 0.8341145091415176 0.8396152032949108 0.8071791857301006 0.749259925623849 0.6834393639558736 0.6090282364047113 0.5340801952077282 0.4745736484141307 0.4497804315121435 0.472450827330014 0.5478020300104459 0.6725100986608693 0.8391377474269629 1.030738090542012 1.2139406127730559 1.3605472203212379 1.4608519579460908 1.5147461066195997 1.5151063750327747 1.4458928276736853 1.2999192737473313 1.0903196109710007 0.8350958779801277 0.5552367107657843 0.27868202629191885 0.03523520749881175 -0.15940010632166954 -0.2984345078183025 -0.3851331899495692 -0.41819175944510234 -0.40467384014062896 -0.3569798341651564 -0.28494381588733914 -0.19369447580251006 -0.09805718457264323 -0.023364608437922124 0.01420512446995673 0.03125406376977949 0.04877036312146823 0.062059637271896895 0.05501318078904362 0.029890740877758875 0.007858047340447639; 0.20066674836803966 0.40104109239907654 0.5873741668122157 0.7444583798582717 0.8531618002214337 0.908043755246173 0.9189806986719716 0.8900221069711406 0.8327822853074814 0.7655784384234868 0.6890971786494966 0.615063812187329 0.561236962130285 0.5474054659750252 0.5852420383497414 0.6747391525144492 0.8099521665168571 0.984042040697186 1.1804203144798837 1.3686200113044669 1.5233877119977175 1.632673324754094 1.6898335711492287 1.684523921716817 1.5955262153968015 1.4173407733924384 1.1687248791693494 0.8688544135303674 0.5410090514617409 0.22152764315598958 -0.057702922783241004 -0.2829392376913925 -0.4511768940534909 -0.5671443620227862 -0.6236546898011045 -0.6216862124729857 -0.5749305627241839 -0.4952837904122178 -0.39194989315079176 -0.28288017850296476 -0.19152013409666216 -0.13522144184974497 -0.0977706776427982 -0.05608489706126985 -0.01720000539441763 -0.0017222709656934683 -0.00799095039950016 -0.01208012320080421; 0.21203770128135316 0.4253092798959953 0.6228786740679872 0.7892285007828469 0.9060455231841504 0.9659745732619316 0.98043462425194 0.9547820294576559 0.8993315406313248 0.8286534376765047 0.747866460505058 0.6723090309557802 0.6204181595492497 0.6125233805922302 0.6624501758156397 0.765636651236314 0.9122303049137304 1.0969761409232985 1.2990598312231267 1.4895410915278742 1.6485424471344998 1.763400593893932 1.8224205385431325 1.807886186433704 1.6994134593960128 1.4943346300653721 1.2133556163366377 0.8751503608067736 0.5054584130560965 0.14839075899348167 -0.1621567724739608 -0.41405718053567886 -0.608150595213415 -0.7515315417583773 -0.8310105031971482 -0.8425948968254681 -0.7999199208830048 -0.7172569556750678 -0.6083859794822385 -0.49314997518920084 -0.3909405014226646 -0.3159425483640898 -0.2543037211384459 -0.1863361945723788 -0.11865803461191754 -0.07297956038994083 -0.05179606316323251 -0.03258755242838501; 0.2269880970702928 0.4507452150069425 0.6531770445706171 0.8192759905962103 0.9360933541114798 0.9998655765197896 1.0193085732276552 1.0006784530775248 0.9521861893765552 0.8830713693180369 0.7980328514613839 0.7167757251190003 0.6594752634990142 0.648700247067533 0.7014129530931882 0.8128939847909573 0.9714952787789358 1.172099071362534 1.3872852761692445 1.5825148304936625 1.7389301798177659 1.8482857640946955 1.8985551819352842 1.869439180317817 1.744246000899542 1.5217927110612546 1.2209865251863787 0.8593226760627891 0.46362564909684983 0.0813027266654698 -0.25205570796873744 -0.5232065122604864 -0.7370769755861221 -0.9019643350472255 -1.001230380750802 -1.027625138427754 -0.9946297040866334 -0.9177476585203437 -0.8134457088802697 -0.6992736256001479 -0.5905829164277911 -0.4973998125333571 -0.4108613927817349 -0.3185330959140506 -0.2249182807656654 -0.14974020260843912 -0.09985792937394969 -0.05496940948350193; 0.24020590493494112 0.4706014472674195 0.6731814577047461 0.832601505576844 0.9446845096821598 1.011692004112403 1.038652695454915 1.031125090440566 0.9924096965568281 0.9255581817150443 0.8359556785735764 0.7475812524877115 0.6842172097244071 0.6693046331313276 0.7203490013992959 0.8345448786555972 1.004824499295779 1.219965615240487 1.4459476444529515 1.6413140497427248 1.7874331314539749 1.8814625900824438 1.9136359544151165 1.8671258412609333 1.729402671643084 1.498171677342343 1.1904505783940433 0.8228791781818453 0.4203564218198092 0.028546765475160395 -0.3153122883872628 -0.5952478159659937 -0.8197338054045378 -0.9984733551242997 -1.1141163548257982 -1.155379706907037 -1.1350998173158195 -1.0726189453388812 -0.9826346074453323 -0.8747022014407928 -0.7613709235915159 -0.6528035113000996 -0.5449812889755504 -0.43160255425015737 -0.3170017507742071 -0.2172411992687594 -0.14177567858742612 -0.07392539976260462; 0.24745764788567537 0.48062334062953777 0.6809613918080026 0.8345402815240469 0.9441010412098371 1.015706845031097 1.0520246688487238 1.0535389364695413 1.0197503162587478 0.9526412875915529 0.8613054414432851 0.7697414609328049 0.7037770282184715 0.6856652370238738 0.7308488087160615 0.8430603670886481 1.018041624574052 1.2363260075258147 1.4597960424139964 1.6458013625387002 1.7757149603002478 1.8520331936304035 1.8681710270092242 1.810108198719666 1.6663803873164087 1.4333050848350877 1.1281874874422249 0.7688563947570477 0.3756507197910725 -0.008652678698300671 -0.34756099456042167 -0.6236457705807389 -0.8473721634244598 -1.0290503145279954 -1.1528895435579194 -1.2070222439536562 -1.2036272765541345 -1.1645785846708958 -1.0988740185457142 -1.0042427384967378 -0.8886220276115352 -0.7682574207447215 -0.6459940634887973 -0.5186484544399163 -0.3877902839484789 -0.26746183426785686 -0.16967130490757515 -0.08463135794684233; 0.2523752604586315 0.4874954938251725 0.6868367109381022 0.8405755800267121 0.9520983971364132 1.026958159155006 1.0654362434858524 1.0645111545929091 1.0261504685562903 0.9557164768879566 0.866787628679217 0.7787859384280945 0.714024400307354 0.6922098627940854 0.7285715725951706 0.832110609077286 0.9987436107205914 1.2030971457408621 1.4085228169954236 1.576059367149562 1.6886010504129538 1.7523421974674749 1.7627728506120803 1.704812167444125 1.5646188281289604 1.337959956422552 1.0449397193361527 0.7055402299229451 0.33630761422675953 -0.024889435191334536 -0.3430860855547015 -0.6044780374797987 -0.8194623014085656 -0.997039679044038 -1.1233047471692215 -1.189679787595759 -1.2053783643498235 -1.1915973700856632 -1.1534617130954887 -1.0782083163357579 -0.9680932000382048 -0.8439033380890958 -0.7128224800048334 -0.5746045949200372 -0.4313180884416163 -0.2960891158535133 -0.18147219758155042 -0.08575540793424134; 0.2600532556907248 0.5001059518431256 0.701457660297811 0.8580337441849573 0.9705584241959497 1.0405797611744165 1.0672942305009763 1.0538999934374185 1.0073258745373643 0.9361321996032861 0.8535163417340261 0.7709309488871936 0.7042103934676758 0.6770928906187808 0.7045372690892586 0.7921513479662338 0.9358343741777247 1.1096330039432767 1.2821843498021537 1.4229204093483219 1.518046800212621 1.5766723920683368 1.593233724262754 1.5506310682489568 1.4299020682106112 1.2260010114676847 0.9583440276648134 0.650136412426943 0.31828548200401086 -0.006883819752326106 -0.29527070990358384 -0.5389651509146822 -0.745074330754205 -0.9155691069830615 -1.0395700067298557 -1.1123137316551814 -1.140141408459053 -1.1431638359249439 -1.1291631544152707 -1.0798447901130022 -0.9912080123148599 -0.8788801695292329 -0.7485997943321357 -0.6047874186542384 -0.45320281910106414 -0.30675648008700096 -0.181585235253248 -0.0822952841097145; 0.2732105980036449 0.5205947604671513 0.7227942198987579 0.8771391916734689 0.9831368657762748 1.0385528054854183 1.0442994593269963 1.012615542240801 0.9576399238011091 0.888818894483527 0.8137073254200271 0.7336419466759606 0.6628677801390922 0.6293081461019768 0.6465772477723755 0.7143649117323257 0.8285228435894527 0.9647113968635364 1.0977332043275174 1.2075069825870979 1.2849450524375732 1.341008452669209 1.3708682271201846 1.357283094728669 1.276637033110952 1.1181333094489032 0.8943015792903419 0.6286121232557177 0.3403696425239063 0.05348464533559039 -0.20793853621888714 -0.4376820299046751 -0.6352336203355979 -0.7952951581829811 -0.9102395176250482 -0.9803628761446402 -1.0130295304629493 -1.0273193041059772 -1.0319774236083128 -1.008441718574366 -0.9492021419041986 -0.8601305607890721 -0.7435196228664144 -0.6063893861169762 -0.45579199821169913 -0.30490868817625094 -0.17598382536715132 -0.0777099083132041; 0.2897356661175578 0.5473626066462737 0.750771980922883 0.9000406958579293 0.99449515275256 1.0282263254617505 1.0054329319070352 0.9492813871410257 0.882379199136203 0.8109556090336564 0.7372967800466752 0.657584892532622 0.5845674039856175 0.5462105447133648 0.5527703266354002 0.6010559398993085 0.6846785227991733 0.7840197285467748 0.8787922974058104 0.9587150939663233 1.0184092361129096 1.071202206852692 1.1178233631140262 1.144008222084117 1.1243007726352097 1.0365397137912056 0.8792081424728938 0.6665072867568225 0.4180145341701932 0.1587550336744191 -0.08779055602399377 -0.310056181488616 -0.4990936005271469 -0.6487370466306531 -0.7556005803568665 -0.8232093101410579 -0.8604125211699267 -0.8837201680054306 -0.8987031881973392 -0.8913312900899302 -0.8554490662074938 -0.7893842122392823 -0.6927469414038163 -0.5733124916911015 -0.43411516032561276 -0.29015846537985673 -0.16738979368933438 -0.07442167953676385; 0.30428312079156533 0.573648024734798 0.7839863760056122 0.9351774023491711 1.022587079790517 1.0341666828794498 0.9782621682815275 0.8892025793525311 0.7972831517552152 0.7070624643110085 0.6214981684260518 0.538291194332605 0.4658368851251656 0.4273271336761199 0.4270097940700107 0.458939774189711 0.5166206613359983 0.5845976944741007 0.648119355685435 0.7034262994700478 0.7493136464776203 0.8018781729132651 0.8702299714560348 0.9454118105105371 1.0009641066926016 0.999565645179918 0.9210008390313049 0.7619622626195061 0.5419666305403541 0.29505060929109794 0.05294676287122044 -0.1641047467624552 -0.34246378311511383 -0.4808769100034731 -0.5827639013895226 -0.6532185768219245 -0.7001509219109232 -0.734560436678524 -0.7565107608494771 -0.7555281901719587 -0.7315125772814944 -0.6822078806522219 -0.6057691724283503 -0.5083246926860332 -0.3908225099161868 -0.26717469906481545 -0.1601156452498267 -0.07392353185754422; 0.31317574425344086 0.5920460145194694 0.8129069627154722 0.9718397145005224 1.0588124946048232 1.056773616745351 0.972800200328874 0.8447506157595563 0.7110047175908029 0.5855130130470946 0.47323383704635774 0.37997581236073374 0.3093305965360581 0.2731652526708331 0.26706109353531904 0.2843442209088753 0.32103817135233625 0.366568308900666 0.4111984719631688 0.4525522931144074 0.49584907044202675 0.5596005625612447 0.658986851876473 0.7911002472875507 0.9271313401962449 1.0139485796474474 1.008710580514286 0.8909583745056187 0.6854655403173956 0.4427726434083275 0.204113442040026 -0.005394648677599143 -0.1725758693506014 -0.30144540800242325 -0.4002368039381114 -0.47462235406613645 -0.5321857157520287 -0.5760007815273842 -0.6018506332811265 -0.6035190392794646 -0.5864173041591516 -0.5520962908830833 -0.49793030764907975 -0.4258448092677657 -0.3367025284339979 -0.24083775503604182 -0.15387725335247293 -0.07442266730851144; 0.31474239031670215 0.5964976758590538 0.8255732314069452 0.992582390973096 1.0813450121315649 1.0692643565531361 0.9616669366806 0.7941213873553666 0.6129793513082532 0.44452673917858204 0.29989298304192474 0.1918817939818532 0.12095430776555684 0.08518420578710634 0.07319085742111356 0.07734395030571993 0.09857330327240395 0.1332500494260123 0.1730260514947934 0.21452595293629187 0.2689932798087573 0.3560165228743097 0.49402146331469693 0.6832142275892423 0.8921645780646232 1.0563176558522684 1.1109633973707749 1.0241999634184584 0.829277817389202 0.5941726268530774 0.3640931038472579 0.16339281308051637 0.0038429824097336454 -0.12069815113550639 -0.21791876256563122 -0.29142858787449927 -0.3496434213613979 -0.39329429157252715 -0.41813497192977056 -0.42232798762306967 -0.414845370993351 -0.3992159421350376 -0.3709658035612827 -0.32834092498764506 -0.2715391717197991 -0.20530159201371262 -0.13909220436615494 -0.06946670073815754; 0.3116908338383549 0.5899240286890532 0.8196808282664979 0.9888663939134181 1.076654356256183 1.0552086645984597 0.9274598244793493 0.7269265479336801 0.5056847085898513 0.2988832165489796 0.1231268643446525 -0.004139886172336987 -0.08301240744996898 -0.12513700546773582 -0.14669952629360503 -0.15221437663336995 -0.1359812334782295 -0.09838227115647027 -0.04889122309139184 0.008518683792221295 0.0831645233412847 0.19657076722808703 0.37048721629778963 0.6034884443935066 0.8611171513615132 1.0771814365201597 1.1789380406349879 1.1276413219420838 0.9578240735298118 0.7433174532745247 0.5304950292093219 0.3421887623456822 0.1881437030079003 0.06382097420682392 -0.03304368661080486 -0.10259827511611772 -0.15180286844238638 -0.18603010383313467 -0.2065715293542107 -0.21393702722404387 -0.2177486258389366 -0.22279269446435176 -0.22311820935299084 -0.21373309357596104 -0.19018136190302842 -0.15323071760604523 -0.10820842769720264 -0.05523372590424404; 0.308708308692218 0.5788735814471291 0.7983848389210193 0.9588478368909747 1.0378404110582695 1.0042958532810182 0.8600511647938991 0.6378707498503267 0.39010780733586825 0.15475703470687022 -0.04921830308891566 -0.20041900812974994 -0.29541174467043163 -0.34981039924198304 -0.378218563439756 -0.38300113715574235 -0.3575042025879996 -0.3028115739024069 -0.2296738445385273 -0.14572700162492544 -0.04658066481448362 0.09196288005699438 0.2918634840035892 0.5474873757575638 0.821566134683544 1.0557042321396983 1.1855388039131356 1.1772764663697768 1.056634064549885 0.8856569399233222 0.7064804911366511 0.5416912821106262 0.39787249597649504 0.2737332420874411 0.1759523291297765 0.10858974940801645 0.06447167975348088 0.03457991031664227 0.013349638352556441 -0.0009381476756278562 -0.01715730111426026 -0.04053252865685856 -0.06803166076373832 -0.09065476402341902 -0.0987402368033618 -0.08986696182225115 -0.06753568671040747 -0.03577634491527682; 0.30148872463646587 0.5614272233705229 0.7648413368696011 0.908300686915924 0.9698970347468144 0.9205921619681603 0.7614092212238456 0.5242128330691779 0.2561673043720638 -0.004828498683600645 -0.23700664991769999 -0.41303934704351386 -0.5237190753859066 -0.5835629414309353 -0.6054130845094544 -0.5938665333577499 -0.5443696669463224 -0.46062173343556184 -0.35585570819934204 -0.23909606300687336 -0.10739271867023402 0.05701949427746135 0.2713416250532549 0.5275721752611897 0.791872519442839 1.0210099010281157 1.167279621746697 1.2010912689243225 1.1387507350993065 1.0266185382733473 0.8989970048061194 0.7740262994171043 0.6523547796800152 0.5345838259159111 0.4344402178683602 0.36071499131214674 0.3069217602352998 0.26507904439852453 0.2307676748497435 0.20238288527540854 0.17246229888485728 0.1328246212040807 0.08142514972041784 0.02937549175625686 -0.008587539298624413 -0.026382384175520582 -0.027235725197626875 -0.016820138768919555; 0.28453446089802703 0.5285864107512028 0.7120708926844157 0.8328582803029297 0.870541356044374 0.8020343257254937 0.6291721667771645 0.3779082478729895 0.09033719185381753 -0.1981406240626531 -0.45740799421299627 -0.6547558093761615 -0.7735965254414969 -0.8242544168687467 -0.821365939525093 -0.775186165755206 -0.6868526155173446 -0.5649676817544808 -0.4238525629514863 -0.267851007661348 -0.09566523059418448 0.09623015151282824 0.31537986187017114 0.5535898254945593 0.7915776394987578 1.0053238898348453 1.1606867332184296 1.2319462969661514 1.2249970076968062 1.1733411198830097 1.1040861277853458 1.0288662482243183 0.9415480860505822 0.8408089054086502 0.7422182505461575 0.6580378569602022 0.5839211775113495 0.5154685833147602 0.4540178483307702 0.4009413100787779 0.34974850606820895 0.288015566585365 0.2112398815829638 0.1303002476803752 0.06480136821298856 0.024973356193701095 0.005667010048501968 -0.0012976278669376052; 0.2557804485015713 0.47543571733146617 0.6348861760824472 0.7287750871946502 0.7387835749046364 0.6483173098147192 0.4604691051710262 0.19365947638649092 -0.11454345807528733 -0.42835083183802714 -0.7089903421636586 -0.9183797125960738 -1.035144350715437 -1.0625927432909616 -1.018066838308219 -0.9213284073977211 -0.7842150724211037 -0.6192618864510109 -0.43881034732439905 -0.24155109821411955 -0.028563358264520722 0.18915954798868803 0.4059862648865496 0.6177643470663685 0.8246476492188947 1.0200842547526237 1.1778898328219503 1.275941251152753 1.3141314368371355 1.3161979941399344 1.3017271209802972 1.2770044755223722 1.2320118465129895 1.159394803660957 1.0704758272060644 0.9792515650567037 0.8842879397906607 0.783207231000417 0.6832039865725523 0.5922137829931452 0.5080880238203982 0.4155141101140108 0.3096539356966786 0.20136880828415674 0.11324847902236967 0.05656678996610149 0.024451374129262623 0.006697528904373199; 0.21997443740804484 0.4074289185566199 0.5378301673678932 0.6019017493558656 0.5836216803772989 0.47004287904198194 0.26353937519824394 -0.020104881622068714 -0.34597906986017957 -0.6753699193314566 -0.9663127063394968 -1.175284679476039 -1.2780377750240095 -1.2726869551613536 -1.1777123007044796 -1.0237219572836969 -0.8339227487991022 -0.6250480971086916 -0.40666484465630076 -0.17319521363941684 0.07211343162422829 0.3069306406872845 0.5164723967172787 0.7039795308716877 0.8815247405356761 1.0548081755084024 1.2082224674372863 1.3259366198315197 1.4021446132516053 1.4514051728133355 1.4847430340160688 1.5018838497952272 1.4926225498463663 1.448846197356315 1.3759025516216095 1.2843637337831546 1.174380665089464 1.0434719531561023 0.9010496632682217 0.7627452919961452 0.6337348817004609 0.5019970926743289 0.3660544308008899 0.2379521883909755 0.13648332776218153 0.06948005873665891 0.02898760684243141 0.007352661381543478; 0.18049161184522564 0.3307400125294982 0.4296424822900886 0.4638899720829138 0.4170046848554241 0.27978137230396183 0.053702514090566686 -0.2433353333740449 -0.5779359994233052 -0.9108474072753616 -1.2003327998700668 -1.3981975200819454 -1.47972415087839 -1.440016153780109 -1.295857092795582 -1.0851421819096965 -0.8398624849921175 -0.5839132168164577 -0.3282312898994144 -0.06816984971773793 0.19276420698971317 0.43045924108851086 0.6288529875436024 0.7942221106222881 0.9456879447983595 1.0953131420906086 1.2386405795478406 1.3707673850788493 1.4817732239304355 1.5732190187990005 1.6469231333324057 1.6966389334106557 1.7118576416347886 1.6880492440654078 1.6288969073906614 1.5398091808574303 1.4184474952437611 1.2612015671690502 1.0772069493111056 0.8891964815599922 0.7094273952735803 0.535042847987852 0.37375974573632964 0.2381420175976718 0.13623248374028155 0.06733079439490824 0.024264241523706712 0.0042598883921659385; 0.13593540257231504 0.24393094686575523 0.3060795904715408 0.3055502063763769 0.22799520119925118 0.07071883379250322 -0.16505554023033528 -0.4648054452913531 -0.7980436633094651 -1.1265010064043277 -1.403788450343058 -1.5789862549265035 -1.630258874670669 -1.5548127176337778 -1.3666820533200603 -1.104420390102205 -0.8055254934351302 -0.5021566970062814 -0.21230892754465747 0.06600082965459524 0.3294063934261113 0.5577325226695207 0.7392106424731831 0.882907339490559 1.0085137607622603 1.1326727632546072 1.2631336652452405 1.404979176221387 1.5462951457072416 1.6748630729001233 1.7826788667256523 1.858622426129689 1.8917621182923752 1.8819254760556323 1.8327128193686064 1.7424074922771726 1.6044702290118635 1.4182333303354602 1.1944946238598382 0.9595347157785229 0.7325992406305057 0.5201970463142847 0.3392758763201728 0.2024002766665321 0.10698275458137005 0.0411940200968382 0.002422445168675967 -0.008164449890100666; 0.08385406606349106 0.1431411538253276 0.16303209055236845 0.12483970723432464 0.017122916424215425 -0.15524798674260532 -0.3902673244911392 -0.6807334895780227 -1.0022611759983924 -1.315380364592804 -1.5666217216908092 -1.705827057245661 -1.717414083519974 -1.604600754285135 -1.3780049918864974 -1.074952914144542 -0.7367433948665773 -0.39764845820012334 -0.07917795522756765 0.21367367150888994 0.4748520365675091 0.6890047635072076 0.8490934736926877 0.9676974970548037 1.066336752040762 1.1667720085276954 1.284452054014401 1.4306316701019925 1.5954543668385783 1.7536958652625818 1.887293235527346 1.9831204907502071 2.0314120031935103 2.0339883116944173 1.9899703904532058 1.8924129023381577 1.7343646227784295 1.5208583712195833 1.2655234043365058 0.9951640595105385 0.7319128433470485 0.48891200412887753 0.2880673043871023 0.14446880750671778 0.05182464232673978 -0.008785114438591668 -0.038028366080548964 -0.03192108671031779; 0.02487408057075069 0.03150254194614655 0.0075313504033574915 -0.06579349856689615 -0.19805264176745882 -0.3773284448384201 -0.6023593029344367 -0.8717739813230498 -1.1687060036683616 -1.4527026365144504 -1.6658707560648665 -1.7607629667235045 -1.7270620834368955 -1.5751618308008153 -1.3170350507361248 -0.988627983713394 -0.63070979331493 -0.27565024009351147 0.05786050218463522 0.35939656245218327 0.6161844809767539 0.8162185905071667 0.9556599942033016 1.0485882940704672 1.1201915399549385 1.1984455387793123 1.3047238911591132 1.4557078592988677 1.6390072995756635 1.8175169543399203 1.967041648668365 2.074481733598019 2.131525814943559 2.1403312232952767 2.0952846380840766 1.9861888086815338 1.80925540466866 1.5752752264416447 1.3017339073881842 1.0130278430376967 0.7283057294767883 0.46328781285626014 0.24054224817158718 0.08071167121734901 -0.017700683630382717 -0.07314434396064681 -0.09000921902038543 -0.06244057533772213; -0.031068228498639126 -0.07242044339635113 -0.1353825043174518 -0.23708232670295332 -0.38455610248380573 -0.5640414411082297 -0.7744540170872082 -1.0163512814643763 -1.2767828799145318 -1.51743378794093 -1.6815563838285392 -1.727320396991586 -1.6470479490161511 -1.4572376556432023 -1.1753391130025201 -0.8364888146087903 -0.4774869640345505 -0.1273962767039235 0.2001813246063916 0.49595723608668424 0.7439824398499404 0.9313860747521422 1.0561632689253317 1.1316596187057386 1.1832432735192044 1.2428583577954597 1.3408446779117793 1.4971772311344596 1.69216594717542 1.8816019041376468 2.037694109537794 2.147492573325409 2.2036645584423415 2.2065138901692234 2.150786623917122 2.0285074697294094 1.8362546472004395 1.5881256955130927 1.3059507716503238 1.011727433754297 0.7175216693843774 0.43888000188526033 0.19696276988521763 0.01935063819159208 -0.08794269252573257 -0.1390115140141042 -0.1420746805426034 -0.09204226385494586; -0.0799336845422008 -0.16058372245816313 -0.25180015692019503 -0.3712110672580901 -0.5237308652564536 -0.6989737599893477 -0.8924323059097838 -1.1036614987367355 -1.3209198935225495 -1.5080953735741705 -1.6146565526179077 -1.6076624894318232 -1.4815785076675343 -1.2583197804416022 -0.9627541089612686 -0.627413175019425 -0.28581269736779513 0.038984695548062595 0.3385784509363245 0.6115708702428493 0.8439680827732434 1.0218766579275935 1.1421578879811993 1.2164446172241938 1.2663085279218669 1.3212458423855848 1.413408236194487 1.5646765966171683 1.754992309664196 1.9421176328310266 2.0964510703596733 2.204610752993006 2.256927235910617 2.2476178900405053 2.173889113688993 2.0349958915838653 1.8295666083800262 1.5705125954757824 1.2826384509506843 0.9863187012724669 0.6889411876342644 0.40485830724262917 0.15581771138193087 -0.029453037180322612 -0.14147401192370263 -0.1903831869508609 -0.18221680855611033 -0.11434303589567715; -0.1261919262647385 -0.24064750222739528 -0.3486824574716578 -0.47129851386005106 -0.617657669689772 -0.7845783936479891 -0.9618042337057285 -1.1420465108502538 -1.3132416456257066 -1.4404052655253516 -1.4831277768594 -1.421284377725513 -1.2545407960708106 -1.0067140675043784 -0.7076143326604303 -0.38778179852038547 -0.07712916011316831 0.20726914015453707 0.46362613806068836 0.6995324509434478 0.9058868091094369 1.0704329973176812 1.189393325401079 1.2703360433124187 1.3296530074514568 1.3898071992954049 1.4768178267392664 1.6142274411297945 1.7881678851217464 1.966002572015339 2.118548313618303 2.2283251528593278 2.279728550641538 2.2633607477759083 2.17683567385132 2.024812049824504 1.8090623630627443 1.5396075902842503 1.2404020184207711 0.9369058234738715 0.6400959564619829 0.36228668068540537 0.1219417374135179 -0.057160996918070854 -0.1680738192901522 -0.2157016737864477 -0.20180671450623336 -0.12458798078407168; -0.16734594165507072 -0.31049347087697265 -0.42935195315039526 -0.5455071422336527 -0.6770387506510598 -0.8288078665720495 -0.98779983576365 -1.1371423684674096 -1.261879321550021 -1.3295206117378444 -1.3121892772576482 -1.2002398931633351 -1.0012918681325695 -0.7387678637366218 -0.4429087652670136 -0.14420558073969286 0.1283439727062784 0.36430409761833576 0.5695681824356945 0.7555788130116878 0.9206935808133941 1.0595054215284136 1.1697640563339184 1.254442500415316 1.3221378572591875 1.3873970700786136 1.4686795419159933 1.5903693369667278 1.7472264334984609 1.9165706743861792 2.070546019739245 2.1879633200998314 2.2487009171621546 2.2388950944620207 2.154635899953814 2.002052071969609 1.782925082812088 1.5049786151291769 1.1924380314555276 0.8790937723160528 0.5854713574190601 0.32072639887990617 0.09716522122774647 -0.0700875032641203 -0.1768713557537912 -0.22219078145633478 -0.2055240999705643 -0.12553919628549562; -0.19938251037740923 -0.36549422698616757 -0.49042825251013017 -0.5931309343062997 -0.7003385047407742 -0.8266816420826655 -0.960486774446763 -1.0806904542530305 -1.1657322703012933 -1.1850714530451323 -1.1205469462253694 -0.9710413750588247 -0.7502562832348046 -0.48378011826125145 -0.201329582785157 0.06909434838744054 0.29921236761835157 0.4842497673840951 0.6364834901133382 0.7656911276623998 0.8785083577456125 0.9790768874000363 1.0704068149490464 1.1522448272688892 1.2240760558730643 1.2910733994594545 1.3681814568710444 1.476970241745646 1.6187971967734323 1.77831622742713 1.9343626501368485 2.0646195500935276 2.1455538331142074 2.1567882924757997 2.0903127316079346 1.9506368231359976 1.7385136423073715 1.4630608513210035 1.1472357867615437 0.8307377902475559 0.5424467230679559 0.29269533571687295 0.0876141617135912 -0.06670460012914943 -0.16769695148981714 -0.21101289572804635 -0.1952759269120635 -0.1191382488190383; -0.21362803232254174 -0.3915006369561937 -0.518610198283944 -0.6085451254483337 -0.6886162334029358 -0.781007686477026 -0.8821126039016434 -0.9722587374176205 -1.025943581524316 -1.0144313761861536 -0.9230164923448908 -0.7536234161163667 -0.5233361065659506 -0.2617899648076814 -0.001585177293534679 0.2333232698258123 0.4188031377455466 0.5548409994420122 0.6557903070601097 0.7295287450690109 0.7870227897808098 0.8421595911651162 0.905176931790449 0.9742782760199445 1.0426284421775793 1.1077039156010964 1.1822304091216524 1.2826259734470455 1.4127071473645811 1.5629862189724237 1.7220959826209663 1.8695618810458507 1.9786446562057693 2.019312774707032 1.9784323841945446 1.8592962523767036 1.665659791159182 1.4071071006525422 1.1040152171177517 0.7966046382539318 0.5189403631478975 0.2824312526353208 0.09132606069320508 -0.05244277036761878 -0.14536470882481123 -0.18547505233616024 -0.17184666172970248 -0.1047061463917257; -0.21317056559182426 -0.39316506222000536 -0.5184716725387417 -0.5953829697518529 -0.6497764063615111 -0.7064936952261053 -0.7726279511998408 -0.8329592016919053 -0.862255409487895 -0.8327929629419077 -0.7315666542415786 -0.5587142643698046 -0.33247227515082195 -0.08420461008547254 0.15082991086066364 0.3479239241385424 0.4885307021088217 0.5770261636158975 0.6290843746620072 0.651972055538641 0.6585809435608375 0.6670262974133523 0.695131364125613 0.7427599791131866 0.7987563263689442 0.8559312531734501 0.9269938653199558 1.0241209345815712 1.1482903481098135 1.2943547082877667 1.4578502922407668 1.6235752789158724 1.7645770539317394 1.8406189732689067 1.8308734362722454 1.7389151741405418 1.5721058295405028 1.3404173429901811 1.0620824786964298 0.7747385280487253 0.5120923818472777 0.28723991598460497 0.10478170172926404 -0.030550760941904835 -0.11517837045616837 -0.14940050264474786 -0.13745275224490164 -0.08309165784223838; -0.20881346851245589 -0.38639846422092156 -0.5082244930875474 -0.5730096593543048 -0.6045433443760243 -0.629050881327847 -0.6633948870999786 -0.6967220459077328 -0.7054441464271923 -0.6608889130937164 -0.5548351695486085 -0.3872681155922901 -0.17429859876803208 0.056301878541396004 0.2660330720179948 0.4250783970046437 0.5188710114177878 0.5584001457801099 0.5631667937383041 0.5408785008839522 0.5043600702745075 0.4700866333984316 0.46127938047430145 0.481863292640154 0.5178658302174028 0.5592486243527053 0.621006968587551 0.7155020373223742 0.8407453896785732 0.9916017789357122 1.1650304471788646 1.3497684281721367 1.5211025845228017 1.634037984769709 1.6594936392227129 1.6010450169922328 1.4691986195066211 1.2719917190768182 1.0274184792358416 0.7686084674962254 0.5257572357111026 0.3133261813868225 0.13879695881282894 0.009898093255204268 -0.0701640731107268 -0.1023190514727775 -0.09502648626915147 -0.05734695919725902; -0.20356800029690297 -0.37669118379029637 -0.49397583229825254 -0.5507621349716276 -0.5664253017294837 -0.569232310492939 -0.5799525787061673 -0.5900897756233829 -0.5774658054130077 -0.5152003818873754 -0.3990937508866802 -0.23456110594798205 -0.03720591499562378 0.17069325635599775 0.35089389569690194 0.4705288386528189 0.5181155897621764 0.5100097208305739 0.4690801819624446 0.40626020899905935 0.3327817757687217 0.2624705681439188 0.22002190931954813 0.20977521567779353 0.2201448030180661 0.24092385659923574 0.28897746615654885 0.37960014043103574 0.5111430925968891 0.6746343030068955 0.865089563914735 1.0719279480722594 1.2708498941367956 1.4177950190343211 1.4783135440604098 1.4573534645719706 1.3652517734526453 1.2074126483368337 1.0023218276137402 0.7798308689849364 0.5631458749140693 0.3668038537084719 0.20030960897019823 0.07443491354914263 -0.007213014672436979 -0.04551529395413135 -0.048501635570823355 -0.029777836713500445; -0.19528985738002366 -0.36203313187436437 -0.4732970220429246 -0.5246269481904094 -0.5340996135288514 -0.5283608164746204 -0.5260730101281 -0.5199127371893608 -0.4885282838273457 -0.40845985842470955 -0.27902465204915505 -0.1122974666070663 0.07176910636788954 0.2531927045578935 0.39969754310861344 0.4811127933052884 0.4886036849064472 0.438541330420349 0.35453617271822035 0.2549344812651705 0.14965173322857286 0.05011623404914545 -0.021471439986458733 -0.0611165929942359 -0.07588876106430088 -0.07422388639546547 -0.037763160638784966 0.052256946029102566 0.19310413617516264 0.3736782090235047 0.585572380259084 0.8133660952354981 1.0317882967981347 1.2023557225202128 1.2919649986440067 1.3064707309804287 1.2549098847937874 1.1405204343933963 0.9815287202631431 0.802195486861125 0.6202918848659498 0.44561540599797894 0.2875014637121715 0.16004811648910375 0.0709460085943138 0.017849923591862178 -0.0045844633409553165 -0.007061827842902266; -0.18472079308806263 -0.34329163283412345 -0.44942943905558835 -0.4986560181122788 -0.5102453688716708 -0.5065063263726157 -0.5007545629806472 -0.4850195639880322 -0.4398392218832118 -0.34522089200780204 -0.2038427058461919 -0.034740451356553556 0.13786354424930788 0.29421660231328395 0.41037541436448177 0.4599279024310807 0.4345834852348993 0.3488020004929823 0.2294391182933254 0.09857552568203043 -0.03368968234557732 -0.15670519300974392 -0.2529315546899461 -0.31572757289899284 -0.34762703988759946 -0.35525758826996706 -0.3232857416263372 -0.2297697632610315 -0.07893361118158214 0.1172627720299357 0.3468662407767608 0.5882341193209172 0.81502101471053 0.9956432810617868 1.1035630811098471 1.1459253650655283 1.1313139991173147 1.0622912943754639 0.9525495778917448 0.8209326391024617 0.6789524528422249 0.5313779195790072 0.38605671080057535 0.2593910991166609 0.16028380085174534 0.08817235242987287 0.04019056760431577 0.013704466882009275; -0.17518457088812264 -0.3276536080383145 -0.43424773903236596 -0.49028699268058445 -0.5101386130079492 -0.5108356068401995 -0.5030966116368891 -0.4795688251579805 -0.4230512780304122 -0.3181027747704359 -0.17044214928965065 -0.004061987276998207 0.1580713377383566 0.2975931746416367 0.39426846746999467 0.4228270814572225 0.3723849313233728 0.25677114040687266 0.10877696477341114 -0.048321541045871615 -0.20399079597620895 -0.3467499052287073 -0.46242390401879885 -0.5409298613410418 -0.5814328250931832 -0.5899281409871869 -0.5554574123859632 -0.45662825955979663 -0.2980768446526025 -0.09309865553381881 0.1457481030151439 0.3926908690373209 0.6196071428640066 0.8001179871780253 0.9167249841111423 0.9771391131594394 0.9932294335343831 0.9678825678391231 0.9065987709309415 0.820830069023133 0.7166700841079883 0.5965518222754355 0.46811021358504346 0.3469784394801635 0.24341367085669813 0.15852189399215744 0.09001610933366919 0.03850591632316134; -0.169792754941379 -0.3217419571896048 -0.4357792028056925 -0.5061851869897899 -0.5359646375377476 -0.5365376426725298 -0.5182586356311006 -0.48261235670393227 -0.41716141398316287 -0.3106061770575144 -0.16654086159317938 -0.006986955685007639 0.14722300918519482 0.2766838285803025 0.36358973320244475 0.38155253366564745 0.3141392982901736 0.17432000250734825 -0.00017808352086527346 -0.1815304111761185 -0.3565673597685923 -0.5105276519613462 -0.6339560772427788 -0.7174950912368838 -0.7578720227305958 -0.7635035004456694 -0.7260263827153917 -0.6270225819908898 -0.4691549731885652 -0.2654363523010491 -0.027416142103110514 0.21808169411586784 0.4397204156510659 0.6131304792234786 0.7315254632697462 0.8056943727926708 0.8477623709416442 0.8590742590524234 0.838277745451919 0.7910473795815551 0.7186571140139967 0.6235483213167565 0.5134694537663901 0.402826443808473 0.30299779723167036 0.21511378638702092 0.13595364576307506 0.06410418223325202; -0.17193948414468035 -0.32604409369538834 -0.4472345836191734 -0.527875660959446 -0.5622397712492786 -0.5563863201651403 -0.5204671005393782 -0.46884029440722197 -0.3989711680840034 -0.30079553549581434 -0.1713822755141904 -0.023661417866804254 0.12386931754856642 0.24674239393428618 0.3266517713358748 0.33749513939034026 0.2591787785224003 0.10191977396076914 -0.09412739190547555 -0.2944177563476078 -0.4808252169733748 -0.6341697419681395 -0.7511605564299271 -0.8274114182793486 -0.8596821426111686 -0.8574049095724725 -0.8164474073956167 -0.7227146690333145 -0.5765699712015339 -0.38646674265550024 -0.1628607019600312 0.06892585687141681 0.2767869127006417 0.43706289992576053 0.5518476629931016 0.6361217830272867 0.6989254744419204 0.7361903272028667 0.7441496314612118 0.725060597544478 0.6789883004418975 0.6072538450791698 0.5165448365126828 0.42088306541680093 0.3334125403687824 0.25191076627553516 0.16939769159659696 0.08388077725389163; -0.1773654488557515 -0.33217163709776304 -0.45218520715161786 -0.5296732124255116 -0.5586652520870387 -0.5418938527576109 -0.4904623256048832 -0.4261280206314861 -0.3563976580988838 -0.27349572487955126 -0.16764509597397845 -0.040701646584626855 0.09433061125939077 0.21010811333802482 0.2837931040663235 0.28850881348069907 0.20379811379848542 0.03811266119696804 -0.16914519071034922 -0.3786372945912682 -0.564890313806834 -0.7076950254204044 -0.8085434238308501 -0.8676949828688006 -0.8854374389121824 -0.8689651945593044 -0.8213575128122508 -0.7339371333471341 -0.6058788408050082 -0.44049607479387726 -0.24574730260698138 -0.043186181958821415 0.1400895107083132 0.2838325323890222 0.39315489085083055 0.48380951185736826 0.5580935417704935 0.606793964528494 0.6283428699909754 0.6261383507510309 0.599841895469696 0.5499715094384147 0.4797378266865502 0.40212579165334245 0.32930450110245807 0.2572995882562503 0.17735200345571558 0.08988280983422035; -0.17527365396833158 -0.3235873438204188 -0.4323527959696036 -0.49605028432007203 -0.5120655902521595 -0.4838839684373964 -0.42436259123348724 -0.35529324233505416 -0.2886146617584754 -0.22107754041210528 -0.13818891605382688 -0.03584004814443468 0.07588154536665691 0.17397660405521248 0.23656628277457814 0.23571831961599296 0.1501491913734552 -0.010558284680941223 -0.21232987799028 -0.41552538839522246 -0.5880042910644832 -0.7122915882092311 -0.7904379610720424 -0.8265596764550127 -0.8268382731281818 -0.7971587821629861 -0.7444489212341631 -0.6666484546650709 -0.5611069505030971 -0.42706677552373823 -0.2709943544530861 -0.11072284128222781 0.03712424438110786 0.15887703999263547 0.258747628048281 0.34970574994200776 0.4271622391256265 0.47842676838353093 0.5045190447841109 0.5112561561647477 0.4986836316888839 0.466313799255263 0.41560267415353974 0.35571420309256924 0.29574774112188934 0.23330667323867765 0.16222510405115648 0.08321715590108508; -0.15704915642158193 -0.28383583620666863 -0.3694546629523583 -0.41309498565849334 -0.4159298472142179 -0.3841387114588648 -0.327840071466261 -0.2637253018649513 -0.20471368063101186 -0.1513027936280338 -0.08833992237382066 -0.011056759585116394 0.07302383381296121 0.1476447602442843 0.19461475528914562 0.18887564383798805 0.1107347362480321 -0.0305138126914014 -0.20876438876260275 -0.387272701447139 -0.532865843672339 -0.6340004243290305 -0.6903263985169439 -0.7062453170403021 -0.6936431115187606 -0.6599256957667244 -0.6085153078843981 -0.5419878133248535 -0.45923526899779366 -0.3593950015658286 -0.24728097310299524 -0.134367117047214 -0.026656559522865827 0.06817653912011142 0.15382618423753538 0.23756270658608944 0.3093195187879999 0.356293555750393 0.3798776055560894 0.388257907688608 0.3841299391227331 0.36591473511490336 0.333690231881742 0.2917286567491367 0.2443817378138808 0.1926107982087456 0.13459957770934117 0.06959072861166651; -0.1211420100254873 -0.21311018624637748 -0.2694408842804928 -0.2938766922834425 -0.28944041522072383 -0.2613304425846643 -0.21747388820156088 -0.16711819847424722 -0.12123974591318469 -0.08169268809122465 -0.037395380526550275 0.014980210614862826 0.0716287459035309 0.1217055691967925 0.15014124861109965 0.140089293822383 0.07911917657528968 -0.026740563436968578 -0.16057860145515626 -0.2951336637381248 -0.4022278600671061 -0.4755702331276026 -0.5133335588080877 -0.5174650086483688 -0.5019746243961503 -0.4726519341914025 -0.4304855242411306 -0.38058703988319537 -0.3243998630189301 -0.26005617388222424 -0.19043600471410205 -0.12111682183460362 -0.052271317654839465 0.012795674311130573 0.07845376196318218 0.14582390632373068 0.20338277679560005 0.23980548914354574 0.25618274913430783 0.2615533224329588 0.2624470775361669 0.25655000674670203 0.23949522676487578 0.2116138572565979 0.17637571863159662 0.13823066558604274 0.09697145361569205 0.05026851520197798; -0.06716788425361586 -0.11616446004005994 -0.14400256663307787 -0.1546734125432943 -0.14997998035151303 -0.13133310243081303 -0.10593699786223648 -0.07787546265253247 -0.052596634484763795 -0.030551303060476653 -0.00640266878731473 0.02084902340748609 0.0495785094557459 0.07408566665741692 0.08536172036484452 0.07654922802346431 0.043433027747960574 -0.01228653728180135 -0.08345207098208728 -0.15650365194491064 -0.21460628872105292 -0.2550795255817335 -0.2754314730832395 -0.2746904215902581 -0.2640058963475149 -0.24578080601303864 -0.2211951630815247 -0.1948448147869205 -0.16832992675475356 -0.13848702298322596 -0.10628878321110728 -0.07405043318692352 -0.04056729580171091 -0.006753917991849055 0.03032100953453579 0.06917685139598251 0.10218932822308605 0.12219234641046896 0.13039789773390661 0.13286006614430898 0.13444110623073383 0.13421652604974962 0.12720503969063202 0.1123368192978921 0.09221546176331061 0.07176001055877392 0.05057272695266765 0.026254234659231494], hyperparams = 95.62268622634633))), instrument = (lgR = [0.12015959380339386, 0.11987290689737741, 0.03375622886248061, 0.1364162262915162, 0.1767038743465052, 0.07625211104244703, 0.06626676581195279, 0.1389309876744938, 0.08696684653800922, 0.022209681895174364, 0.10619521459089054, 0.07522801765569398, 0.034866375713591115, 0.10672024351786144, 0.05439896790154933, 0.04213221914864992, 0.09802547843478299, -0.024592743672618603, -0.09680878245805903, 0.03695114122338155, 0.1584221813761927, 0.0567993226764613, -0.007783348328059781, 0.0023243977612495215, -0.24833154607267083, 0.08656441459947233, 0.07529076629478042, 0.11274801238771782, 0.009826903663616885, -0.21578490050401858, 0.029791279593439288, 0.02260286990751114, 0.08497207067902897, -0.36221665243556406, 0.10395830263116738, 0.04720806491038278, 0.0306570870169622, 0.07202010885962754, -0.2970832322593879, 0.05459798561482855, 0.03089926574912477, 0.11962609939255191, -0.18746907697827264, 0.13182940103245694, 0.060005133634369164, 0.04321100599429057, 0.0897698437111948, -0.17789239663729622, 0.16503240064402075, 0.08417514828530742, 0.05327740917766454, 0.10161052504196755, -0.5218571400901266, 0.03141097382209737, 0.17654534884641296, 0.07109939086056591, 0.033659050640981625, 0.13231195765281642, -0.41484224960883753, 0.013173418407802766, 0.18905642832719868, 0.08614793453501919, 0.11835028344010975, 0.13137985462283933, -0.016484610960276015, -0.21672440861663228, 0.22885747257665554, -0.10584583755253266, 0.08620512776239624, 0.0652162546616284, 0.14518782307999795, 0.13002468379925033, -0.2841153491015078, 0.22348945347611246, -0.09114354598560329, 0.08264069295282177, 0.03753347025334432, 0.1974761773601753, 0.2251162973542438, -0.15162835768732605, 0.08725034273412831, 0.07737463978386097, 0.0546793314919656, 0.23903741283067545, 0.2493601920317472, -0.9579268998032628, 0.16741305003901996, 0.09323380761431421, 0.04652321110975496, 0.21853346416690475, 0.27856268280529983, -0.16147035511325625, 0.1346513745456888, 0.10698892867537117, 0.01513693358800682, 0.1774706618182076, 0.34321198611588605, -0.0945227813759857, 0.2089314815321064, 0.08104878791060759, 0.18403171701865967, 0.3958111101580862, -0.29558887132537937, 0.2291173230575013], lgrat = [0.013057400043783893, 0.03737146372801951, 0.02433113212704536, 0.040942953241850204, 0.016051687663759303, 0.03187054422267621, 0.0029640032973680406, -0.007722674675589386, 0.013304585484094097, 0.02177060569622866, 0.0381289740905177, 0.0076267453078294085, -0.001882542615129623, 0.03660021547681239, 0.04267002095605698, -0.03634577897853575, 0.013494291386293274, 0.04735519260565965, 0.059609332283440675, 0.02175259685180132, -0.007712439605365152, 0.03559186314237601, 0.03722329015750399, 0.019109144256104504, 0.04482846191206839, 0.01787674666242816, 0.013057543728428815, -0.0108887389010859, 0.055397702916650325, 0.05959980545212032, 0.03814410001231594, 0.028580347799898563, 0.02940557851314421, 0.060220249025983996, 0.026898982596260713, -0.0008491809887510665, 0.0019206894895106483, 0.05201986645907302, 0.058078326661315606, 0.007335417085049373, 0.06358626896774196, 0.032999850925819564, 0.049863154430838934, 0.05172175678965867, 0.020335772510872315, -0.03275631789767349, 0.05248562589963408, 0.06664322010257342, 0.017620755238962074, -0.006120814467506909, -0.04755345753978201, 0.042796268656579706, -0.13864141153666465, 0.0411968109322375, 0.05297928944782069, 0.0032027741459445024, -0.0021714473870729184, 0.044938816047560774, -0.11400532653148082, 0.044689973959106136, 0.054835996661308055, 0.0046993357713335186, -0.08862958163584798, 0.05650340944278794, -0.012127933879809451, 0.03873201956504509, 0.04005399074586799, -0.12457111774146076, -0.001588968318125576, -0.0025281901197995614, 0.04403442956219011, 0.025020891256250535, 0.0799395905706705, 0.043598731103540435, -0.06735779037510405, 0.019733297134846704, -0.003169449785639228, 0.024552089626320925, 0.05333447875337747, 0.040135555148387625, 0.019686892235358484, 0.0017081115903934923, -0.06600170380900207, 0.013371506459887451, 0.06427778061773531, 0.0760479303104155, -0.013933979796083873, -0.007881925215420997, 0.03226639141562891, 0.04332074136698085, 0.06996805442067057, 0.05445703253690702, 0.03355188289292142, -0.020738739044765385, -0.025929681774930435, 0.06572623006725246, 0.08104101153798936, 0.06527940269654589, 0.07617310574264637, -0.0016082986833766773, 0.059873764695370014, 0.09287544357115966, 0.03032285897222144, 0.05693930962579938], gpR = [0.0, 2.9177087553554735, 0.0, 0.6479357010960742, 2.812743146452651, 0.0, 0.5026260949335113, 2.709843068275733, 0.0, 0.40635218194847206, 2.4492628395732643, 0.0, 0.42640788276512265, 2.3171319909356614, 0.0, 0.4371439111172373, 2.067916043417808, 0.0, 0.36571796489558106, -0.8211161921489348, 1.9407366429589314, 0.0, 0.3432037904734064, -1.0475997342498444, -0.24870123008821607, 1.6574007301144498, 0.0, 0.2689101077479082, -1.060354451773074, -0.2360643099982507, 0.0, 0.258640021236254, -1.3026478509597117, -0.19738090247321438, 1.2383329747448446, 0.0, 0.154046519219692, -1.359571405578891, -0.23721659767773579, 0.0, 0.25269398164623236, -1.5082205948729572, -0.3049412347835805, 0.795259564614947, 0.0, 0.19834394224725882, -1.6105984996951772, -0.3507707421605012, 0.714349340103758, 0.0, 0.24651442697694945, -1.8861467530284337, -0.6117160959209914, -0.4800367724479689, 0.38905802233810866, 0.0, 0.20795606878296946, -1.9265586030370698, -0.6059737852287549, -0.573839363395477, 0.2654022334306767, 0.0, 0.22141420880956394, -2.1591884292119343, -0.6611515760282819, -0.8127615009597337, 0.017854931351594012, -2.5825875373741787, 0.0, 0.25342796665322087, -2.28493764626244, -0.6778798705952116, -0.9105959790702101, -0.07512843301077865, -2.6573251732035206, 0.0, 0.28574255263807463, -2.778946344249563, -0.653799793381558, -1.9750764391724176, -1.8675867235090498, 0.0, 0.39607183398763324, 2.9310949045281967, -0.6246262505988447, 3.091759623500619, -1.0683776632249753, 0.0, 0.5275419279112459, 2.294613208154286, -0.580301740368124, 2.3316475113541277, -0.09671853355205444, 0.0, 0.6495923041701703, 1.8596791588506163, -0.5665438437438943, 1.911687599698572, 0.2565855508845461, 0.0, 1.68910935120789, -0.4922121345282658, 1.792710169786858, 0.5025717813834383], gprat = [0.0, -0.047099796543951584, 0.0, -0.07913778554490354, -0.04832599846020827, 0.0, 0.057963154920668544, -0.04046158909584262, 0.0, 0.05519990171484165, -0.05587648087457725, 0.0, 0.044187671016016415, -0.0380483713891619, 0.0, -0.06729012635753893, -0.0705743052545451, 0.0, -0.04901363626080878, -0.036510125169412155, -0.04853459120285392, 0.0, 0.015886207412923826, 0.03906977059628164, 0.03586265404962466, -0.06059642610934863, 0.0, 0.0025673574429553514, -0.015192045893844824, 0.03187235349971252, 0.0, -0.05598447502412801, 0.03339411749635045, -0.009242560027158089, -0.05696776744994093, 0.0, 0.020883482233650073, 0.005940837101565547, 0.022862879090349782, 0.0, -0.06197524864149016, -0.042025444080708955, 0.02649200299342852, -0.036651935226566575, 0.0, 0.03474009479148995, -0.027263922575509605, 0.02696278346103525, -0.05852101265713424, 0.0, -0.03998583253005227, -0.021853458270745644, 0.020764684066081434, 0.012034076897416307, -0.042334248534933214, 0.0, -0.011126438750622565, -0.05401073501342041, 0.017520038360888678, 0.055193757557892575, -0.014939591341744883, 0.0, -0.006271081335230988, -0.09277895266093265, 0.012837105282375187, 0.06270312986469392, -0.024178115442460485, -0.6599121649069929, 0.0, 0.035822782243388106, -0.05483901108551948, 0.013963143543728256, 0.05085612372157977, -0.02880409126651862, -0.2498504044332765, 0.0, -0.015600718755732287, -0.07765511621467763, 0.010899727245619594, -0.019436309268359247, 0.2236118919301868, 0.0, 0.029545983001520896, 0.05707150844913766, -0.0018620055611071316, 0.009242462233174892, 0.1515554867049312, 0.0, 0.0555157267225868, 0.17804804164452306, -0.0040343754403242395, 0.012943146277633783, -0.12661206205272874, 0.0, 0.061829763342768844, 0.19716098632235776, -0.009819242383655622, -0.015245128896674016, 0.04572076220954721, 0.0, 0.17285198080201775, -0.009810941927915779, -0.012400730789005911, -0.026207172804871545], dRx = [-0.008609940061830315, -0.05210812875241912, 0.030930864307888536, 0.002642423346309614, 0.0334885304748988, -0.1083662244624674, -0.03300391598567352], dRy = [-0.03772146039677258, 0.0343411549537251, 0.08734328452022828, 0.0033973523183382743, 0.04553046083737433, 0.012482284609238297, 0.016698408522172938], dLx = [0.009037633571012635, 0.020360158896546215, -0.02770928718326479, -0.05578300647528499, -0.004876214228079984, 0.1397084910999681, -0.0409577991078701], dLy = [-0.03358399545808372, 0.03735991083046108, 0.08638920395384807, 0.06209822104081211, 0.0064312158537089634, 0.00843377025032564, 0.07098347989672463])), retcode: MaxIters
u: [1.203423630234993, -1.6344244699545387, -2.5578955186621286, -4.115130226219029, 0.006826322824203886, 0.015192269242944327, 0.023201720402507297, 0.02786792390546406, 0.0307491246660269, 0.035653496720154165, 0.04579369214763096, 0.06337146230597969, 0.08380925512925763, 0.10040851664518387, 0.11714448472640021, 0.14220303158874348, 0.17569246582376255, 0.21276125716730546, 0.24694010170896727, 0.2768084895273114, 0.30399031618087213, 0.33150516639648886, 0.3605232252985234, 0.3891863818422213, 0.41158644808591405, 0.4209716438857698, 0.41291044062942556, 0.387872946496821, 0.34866167589736385, 0.3013393546867594, 0.2544916388614475, 0.21194754464352572, 0.17236495448754607, 0.13726506465885735, 0.11201229677959013, 0.09751960195155153, 0.09216569735269554, 0.0966665307536784, 0.1120476757855325, 0.135334429628833, 0.157828259650853, 0.1689222370420436, 0.1637297966860292, 0.14675394802571592, 0.1269959900947078, 0.11005828778047118, 0.09605494947723146, 0.08268947479901453, 0.06890547824041834, 0.055020865967412326, 0.039949198044908554, 0.021521843384495934, 0.0135790871283861, 0.02954049900648445, 0.043599857499380966, 0.05250208742173838, 0.06103046174547021, 0.07560728366930991, 0.09937473454738467, 0.131131433429367, 0.163218695729739, 0.19327610078727808, 0.23273781641298893, 0.28958268330206416, 0.3593570179733292, 0.4324407926421055, 0.49939088277574023, 0.5580192826832175, 0.611221574122637, 0.6648373754913555, 0.7218635065839243, 0.7786179394326762, 0.8231894788760499, 0.8421110952001318, 0.8270219314937272, 0.7789141237184865, 0.7064247897662692, 0.6207052098731174, 0.5347989648893455, 0.4562003009557809, 0.38304812298496344, 0.314860197963446, 0.25880586939375894, 0.22110918413618652, 0.203489805411342, 0.20571981861618255, 0.22459534938827075, 0.25433807875884046, 0.2857845144293831, 0.3058089690837244, 0.303840089958593, 0.28176831511177747, 0.2516936826236659, 0.2235413133885241, 0.1987124752452483, 0.17322995929522061, 0.14437279127857042, 0.11353587419421352, 0.08072042839840696, 0.0428448868809746, 0.020607387606374684, 0.04372740175030192, 0.06319515343177147, 0.07725461201871102, 0.09401420080228795, 0.12075187827869828, 0.15772960380443452, 0.19786487461769806, 0.23602156245551084, 0.2833781321445059, 0.3554708305874627, 0.45053971132277354, 0.5566360190300867, 0.6604206029970773, 0.7527764649056058, 0.8327715028819704, 0.9056697475754085, 0.9808796911002442, 1.0631733002749602, 1.1447183108989263, 1.2071703036240244, 1.2321217438403789, 1.2090356485576816, 1.1404406376714848, 1.0431145462282185, 0.9349355647039437, 0.8283408608341514, 0.7304623585367664, 0.6389565720934222, 0.548342714155048, 0.46219131809359654, 0.3924064797831678, 0.3478184834781397, 0.3304226328384414, 0.33492830059978795, 0.3526404420372597, 0.37691330344837126, 0.39971235620589696, 0.40852576276909885, 0.3964408552566762, 0.3694178562819236, 0.3379214888842851, 0.3058601420414648, 0.2699347067911083, 0.22628543782081428, 0.17653314856607985, 0.1230650418918244, 0.06422714034601883, 0.02907146328800041, 0.059955806978925254, 0.08611430200340009, 0.10727733647460991, 0.1327078817268878, 0.1693598463570144, 0.2148955256447154, 0.2603370916510269, 0.30944899119702357, 0.38442715784844567, 0.49437267487680153, 0.6242482448551314, 0.7574876745157563, 0.8799009130970912, 0.9838425395393878, 1.071303899576225, 1.1523248051408517, 1.2411040510630533, 1.3411258172573348, 1.4372428230870709, 1.5034640988478443, 1.5207515426769098, 1.4844255535499522, 1.4013419390949273, 1.2933057961840504, 1.1846882818094115, 1.0842039502098806, 0.9912038072818697, 0.9014095876052401, 0.8065670410128466, 0.704655858430763, 0.6070102750958712, 0.5271915990324366, 0.4730972345939503, 0.44626655918754426, 0.4396336912715433, 0.4462770215043465, 0.46286384916398593, 0.48076318106099747, 0.48626353855671733, 0.472863630836704, 0.44488170984137876, 0.40818143243187976, 0.36331736933027114, 0.30770338245554874, 0.24146833853613384, 0.16724894412868221, 0.08619468272249667, 0.040467809514302455, 0.0796966262602317, 0.11350559005106634, 0.1438063878151573, 0.17853552106558315, 0.22332763919086776, 0.2746763246476311, 0.32753005325260737, 0.3961167646005351, 0.5032085797518796, 0.6436020414455225, 0.7940305006729178, 0.9397643863715552, 1.0661411611240381, 1.1666535745048177, 1.2467555297366517, 1.3224743685212659, 1.410506303734095, 1.5091519787096819, 1.595859438009824, 1.6389738121651751, 1.625813272922753, 1.569824803586575, 1.486367549947105, 1.3946479353786447, 1.3163487488500956, 1.251231241342869, 1.1850227046728188, 1.1107085875299203, 1.0246439114447283, 0.9252007054286919, 0.8191954316340933, 0.7179073063157481, 0.6313379938590084, 0.56990192811982, 0.535102272306112, 0.5196887686543797, 0.5206297471000637, 0.5351977363073434, 0.5513809592675373, 0.5533917594106503, 0.5344351320288969, 0.4975211493832204, 0.44635967050987485, 0.3816536565475767, 0.30313136003524405, 0.21140952317288275, 0.10882389090896276, 0.05445182202753945, 0.10213299894179156, 0.14406900948287094, 0.1862450951348585, 0.23375960724615522, 0.28841977857113366, 0.34660902647896613, 0.41025836416992384, 0.4998889484637884, 0.6312379787280817, 0.7861919847432919, 0.9433661430426381, 1.0908242236827352, 1.2085941200945642, 1.292601632905932, 1.3533955488866545, 1.4103550253420736, 1.4747773730191167, 1.539954710433034, 1.5845659273545625, 1.581575723734796, 1.5274089854544932, 1.4529461691270773, 1.385195317848816, 1.3374873953441713, 1.3170230790184596, 1.3079799931623557, 1.2813996581913345, 1.2253134209338592, 1.1464470725498923, 1.0547597391937036, 0.9593751060327572, 0.8657399945114634, 0.7769916256181567, 0.7025617283181583, 0.6508362819238811, 0.6167816918060259, 0.5958056378076533, 0.5900815964548205, 0.5977972253250721, 0.6056919695798934, 0.5981264015971434, 0.5687669294569031, 0.5176922065289498, 0.4466826574552232, 0.3579490064861609, 0.25211124579425015, 0.13071800474752604, 0.0681472384405584, 0.12490570840138683, 0.17666308542497097, 0.23391799985899042, 0.2981445680899823, 0.36563260753449367, 0.43319857761403796, 0.5074282790012116, 0.6109097004259085, 0.7521093080795215, 0.908274389024676, 1.0663290222767088, 1.2080535925779465, 1.3058059234981196, 1.3620090624446772, 1.3947621810489088, 1.4202430729650553, 1.436982418518967, 1.4395709598134943, 1.4231358050066591, 1.3743859331509916, 1.2934319603732876, 1.215555095729388, 1.1774220444106696, 1.1859302273476144, 1.228380405134225, 1.2748216407972361, 1.2867851303324715, 1.243376075134407, 1.1588243691999538, 1.0616871353281558, 0.9771114396020106, 0.9123530166295607, 0.8578437392733748, 0.808011552187418, 0.766382613775524, 0.7290642987572211, 0.6906654165114641, 0.6565879509071558, 0.6378124136633273, 0.6351993109543561, 0.6336150794348663, 0.6166631245799837, 0.5744471557343573, 0.503078411835259, 0.40587083339665825, 0.28729750453139163, 0.14980847935317657, 0.07937713120284347, 0.14567066585969832, 0.2091426757490671, 0.2826057268328527, 0.3641425479167691, 0.4463819291450455, 0.5235782912622828, 0.6026981128973723, 0.7093658590177983, 0.8477057397685115, 0.9985505857776484, 1.155234146619733, 1.2862532589130076, 1.35744511209218, 1.3775977670752164, 1.3748251801989957, 1.358920334167383, 1.3190538413166941, 1.2552203301644926, 1.1804465716622616, 1.09792115209863, 1.0121767991750148, 0.9531286590935721, 0.9559306266549882, 1.0186208092444982, 1.1107399997038663, 1.1961085396321136, 1.2341475893440983, 1.1953311316159883, 1.0921713713775565, 0.9699989870623926, 0.8776940438971739, 0.8383890568312362, 0.8343682864958385, 0.838234974624109, 0.8367783832594275, 0.820495372908458, 0.7826772546713394, 0.7304871850217721, 0.6824822984431262, 0.6563074372091364, 0.6484567440516855, 0.6393150505153986, 0.6101004143799638, 0.5468303295759468, 0.4464879051199548, 0.31661014995507974, 0.16499451141664018, 0.08753742547711328, 0.16235634442339514, 0.23607817366017544, 0.3225116565802086, 0.41974841227300724, 0.5158009694686786, 0.5969934295605294, 0.6725107214544032, 0.7725126752451104, 0.899014247065158, 1.041616477529215, 1.1974691162770985, 1.3227474592110198, 1.370306757677595, 1.3518304463214599, 1.3076844599902457, 1.2494357463523185, 1.169265649595557, 1.064702954275197, 0.9507297833731626, 0.8493291660758366, 0.7761320762476016, 0.7509834119733948, 0.7908859557797996, 0.8839956261837437, 0.995030578908628, 1.091428423346547, 1.1391141564180634, 1.1035788402821285, 0.9843592001827294, 0.8320117774796936, 0.7148755425295157, 0.6791380503620259, 0.7132262330543983, 0.7733388202054129, 0.8276126446907491, 0.8550946462067116, 0.8417624763547709, 0.7932590193399883, 0.7304268662416566, 0.6804200509723188, 0.656152278970134, 0.6444444677629884, 0.6237196079486625, 0.5727306876676502, 0.47680353699838807, 0.3397491083450248, 0.17631277606458484, 0.09237529321251635, 0.1713659820933557, 0.24960297676960227, 0.34497270091874016, 0.45569458833918675, 0.5597781623140076, 0.6369895333376278, 0.7017996503320618, 0.7864065642944691, 0.8944425329680793, 1.0260619135141504, 1.1824135911919387, 1.3081025690167487, 1.3417163213369796, 1.2951701955462782, 1.2143935276304496, 1.1279858706873718, 1.039179244539616, 0.9309715083862464, 0.8058461740496374, 0.7049607502644478, 0.658356080908697, 0.6681299294633705, 0.7210429101705422, 0.7984634993112011, 0.8800091837630319, 0.9505068929490229, 0.991256641995616, 0.966647687330203, 0.8537203686603623, 0.6885460656162392, 0.5469452777406456, 0.4940532065123445, 0.5376816625949532, 0.6334432040351297, 0.7368209327385788, 0.8170182990149711, 0.8483664121913671, 0.8265521445380193, 0.7707369494433677, 0.7103232292262575, 0.6699707221199201, 0.6486574670486794, 0.6276720495970115, 0.5843257801373625, 0.49574142540613114, 0.35661112887010177, 0.18453634245187833, 0.09226630304496262, 0.1687197135285948, 0.24670598200638452, 0.34906443837684203, 0.46864452506460696, 0.5741171989654498, 0.643415175943674, 0.693125371504206, 0.7568394095945254, 0.841921106443105, 0.9555641813051541, 1.100042211215456, 1.2169648168248015, 1.2519314932626409, 1.2074564750265606, 1.1137433606874503, 1.0180160599640369, 0.9400910593954547, 0.8526314967456395, 0.7451570611506622, 0.6675559755887402, 0.6561841322053021, 0.6905167418650411, 0.7287971945165346, 0.7453932165833757, 0.751110250025816, 0.7669911428471927, 0.7916890773626448, 0.7896867087677983, 0.7121944267137578, 0.5621638611732739, 0.408785040294833, 0.3289025789705272, 0.3534833725731474, 0.45561254256849504, 0.587843673329616, 0.7121580692206754, 0.7956813340287301, 0.8199517486916942, 0.7938802841798289, 0.7437378843000393, 0.6986046593497748, 0.6685626713614182, 0.6412879749063296, 0.5962212749433413, 0.5093100080845885, 0.3690370585254175, 0.19116017415693687, 0.08453027414732561, 0.15472226277320947, 0.23202426704684959, 0.33881511905752254, 0.4622767671290292, 0.5672039026730207, 0.6301903646493152, 0.6676685929122369, 0.7119819640432417, 0.7707986207219111, 0.8538153044804033, 0.960465338867985, 1.0486160463935215, 1.0926391766283772, 1.0798534075617876, 1.0050166463287578, 0.9084386594562921, 0.8315060949392629, 0.7622702197205385, 0.6880709842626274, 0.6481295494742145, 0.6683217879409032, 0.7128176218271495, 0.7227222343416234, 0.6654865044966928, 0.5855024487962066, 0.5430198060915046, 0.5539086221756118, 0.5847135095110177, 0.5671325088454019, 0.46059741147303895, 0.31100017058963225, 0.20197938021392717, 0.18848999770666808, 0.26989339365455534, 0.40822127142488734, 0.5604019656720266, 0.6890135690049312, 0.7673921924645091, 0.7908817741378574, 0.7740552149964558, 0.7422957073063986, 0.712253407175895, 0.6781707620189815, 0.6235601730117981, 0.528901442676484, 0.38296962066096435, 0.19871862571758336, 0.07009454209024621, 0.13337652714226916, 0.21065265099722508, 0.31926754358366105, 0.44439212297666436, 0.5516842708670093, 0.6192186904875658, 0.6581043340555699, 0.6893142102416354, 0.7205179088471547, 0.7622007492410157, 0.8109128362173749, 0.8509420169197802, 0.8868022867544297, 0.9006867354358474, 0.8553174771106247, 0.7598730968740739, 0.6655676689380805, 0.5900908271017207, 0.5332673549736934, 0.5203874464081771, 0.5538393217016774, 0.590313982077725, 0.5713203675738556, 0.46990618848246485, 0.34617129980678224, 0.27322261865084047, 0.281953557891554, 0.3469309656706188, 0.4009308296521888, 0.3636958859946866, 0.23613411089635414, 0.10297519571984129, 0.046881998070029615, 0.09197489773696582, 0.2163076772752556, 0.3808708950112116, 0.5437548728253131, 0.6721844216218109, 0.7538245638077945, 0.7899353672971795, 0.7925340923481757, 0.7765886651613981, 0.7406978685507604, 0.673127512212338, 0.5629531584713492, 0.40501817595406, 0.21038173918763645, 0.05266910694861659, 0.10888776361971336, 0.18577260170871665, 0.2940597995054945, 0.42081950429016757, 0.5398677456083333, 0.6315926161550559, 0.6902426585907101, 0.719111416545094, 0.7257129782631208, 0.7216121615995439, 0.7099980628356873, 0.6950817457247204, 0.6952792393638009, 0.6999258329524476, 0.6625157283200945, 0.5676042939310625, 0.4522721031425971, 0.346304932604304, 0.26919885566195495, 0.24867854021810407, 0.2681101265351502, 0.2768227974259524, 0.22852769740330067, 0.11187819802434378, -0.006726585464925429, -0.06469699916141246, -0.036016671828573196, 0.06412954541422489, 0.18450410600944925, 0.22989439266708206, 0.14522368436775787, 0.002160071726137377, -0.08934785484175935, -0.08152437582751218, 0.01774942330445593, 0.18208163367545893, 0.3723109089266389, 0.5474360841284718, 0.6873636396854537, 0.7855346745251076, 0.8389846177792625, 0.8515910753028473, 0.8217179567996276, 0.7424234579998206, 0.6128297009794436, 0.4373047787580802, 0.22712216786391026, 0.03590662702423783, 0.08497807556084686, 0.15914988325134055, 0.26478479293493906, 0.396622699586865, 0.539037450297731, 0.6716159845467693, 0.769770623250572, 0.8136935262429523, 0.7996102370182772, 0.7487808679997673, 0.6883525857195898, 0.6295458742866707, 0.5845692659473709, 0.55109606847871, 0.49179309387795817, 0.38734832777661615, 0.2538978897441361, 0.11189627633931663, -0.007857531026057754, -0.06761786367302361, -0.0833080124629524, -0.11195764966990113, -0.195143701606967, -0.33138649119709934, -0.4420081774667394, -0.46378126986410884, -0.3940079311624724, -0.24949940472736926, -0.07519046936779109, 0.04256895461828131, 0.01762354104784133, -0.1136833097480423, -0.23220867127599637, -0.2611638968115415, -0.18972160909690736, -0.03264665197617288, 0.18057438248501173, 0.40418377740858696, 0.6049768295173878, 0.7680794212868193, 0.8792199752865106, 0.9292543730602488, 0.9119253019307592, 0.8241380107218692, 0.6749585097409412, 0.47848169967980425, 0.2481430048712062, 0.023138712359150514, 0.06446343026391729, 0.13265362670937303, 0.23536929510704116, 0.37493468241193506, 0.5426705050590088, 0.721895934186079, 0.8736069539670319, 0.943369960951732, 0.9091232125228224, 0.8154717710756274, 0.7210366708378707, 0.6372714188122718, 0.5624819962599384, 0.4928729748213067, 0.39905602790817374, 0.2699965721138765, 0.11977783125742053, -0.04205152379940777, -0.19950085120693403, -0.3085813981577385, -0.36564314160991745, -0.4247768269469232, -0.5329439939276132, -0.6876059164895126, -0.8085302368216079, -0.8128689170994022, -0.7056776353678418, -0.5198277240231965, -0.3079822891109692, -0.1482187202682714, -0.1193680289098991, -0.2222345315626292, -0.3604197887139616, -0.43217475793960136, -0.39316621072850466, -0.24777333893965559, -0.01792734854357228, 0.25285548477707204, 0.5168558251335159, 0.7465167577043119, 0.9170497475382302, 1.00705320099643, 1.0053925192680344, 0.9120725432097851, 0.7446127214696151, 0.5259562762158336, 0.2725475689073512, 0.015874095005949416, 0.050029548867745854, 0.11083731398375984, 0.2096385496344737, 0.352359463537373, 0.5355246582701243, 0.7487686511538277, 0.93972353915049, 1.0244623085569526, 0.9717459824667075, 0.8463494342054225, 0.7290230541130825, 0.6378822515149275, 0.5627767291102768, 0.4856145305323624, 0.37016351356601823, 0.21595474811332344, 0.057979569251885, -0.10285358969020941, -0.28181215599426906, -0.4271828844018718, -0.5181718291042546, -0.5924692972352003, -0.6953201987570176, -0.8421925935116668, -0.9664940109264928, -0.9735979143447836, -0.8509903158708265, -0.6441033521150882, -0.41889442007586214, -0.2520975036926584, -0.20184409405466294, -0.28548404297233865, -0.4409524588275786, -0.5590929810559232, -0.5593961711467996, -0.43061730945907684, -0.19516303821900108, 0.10946361787232499, 0.4293852523645549, 0.7225201917120861, 0.9515348708593762, 1.0816057475152372, 1.09634149899779, 0.999202354104725, 0.8151392064784232, 0.574863100576254, 0.29787744535450267, 0.014858123583543173, 0.04448232962291197, 0.09691000889919801, 0.18775339937857624, 0.32510235410061467, 0.504941867767528, 0.7164977619200267, 0.9022316967771872, 0.9746633824047735, 0.9080475991132525, 0.7689320388226207, 0.6406422747161608, 0.5529315060377871, 0.4970076787845413, 0.4423453377867654, 0.3407319749813461, 0.1933010387807384, 0.04898180807032325, -0.09975537410919742, -0.2728040364249931, -0.4217747910934439, -0.5301112283922663, -0.6072304174755204, -0.6834051950208587, -0.8016229451512931, -0.9121400861933076, -0.9240382628621773, -0.8016526503595374, -0.5863548791384571, -0.3642102098627091, -0.21501436535635163, -0.177364885691588, -0.26961358762814847, -0.45358593445563816, -0.617914268426751, -0.6611817124228889, -0.5561622922938257, -0.3279883112651625, -0.011312817535592493, 0.3452715072016443, 0.6913945536948802, 0.9742397000403606, 1.1428938845131396, 1.1742338630525437, 1.0742315375337395, 0.8751841792315408, 0.6160350947239525, 0.3191621043289123, 0.01958540511948564, 0.04740024494798556, 0.0907006617612805, 0.1695637285362254, 0.29241737574226473, 0.4486501852881677, 0.6216255176892206, 0.7564814592804374, 0.7847063854003482, 0.7007059393297431, 0.562468542132457, 0.4389315674029156, 0.3643425125940691, 0.3339750326364506, 0.3187470717488993, 0.26440235593200956, 0.1587220922438818, 0.048878797991344924, -0.06550002349331034, -0.18897406441275863, -0.29950145397316214, -0.38638092283181036, -0.44342304491398116, -0.4937333778000045, -0.5824710066848623, -0.6841130109650683, -0.7138954964017565, -0.6152721400426946, -0.40550681106030023, -0.18820046186006362, -0.06516907683081759, -0.06085838572459018, -0.17894966648214114, -0.398705557393492, -0.6075324513306049, -0.6941517067803156, -0.6220147997743734, -0.41589392010195586, -0.10872924884952566, 0.2618938371429591, 0.6442859954489036, 0.970899557598982, 1.1742585500323703, 1.2227153968805178, 1.12240802032513, 0.9116870525042439, 0.6388795338703915, 0.330341313336052, 0.027366790812396576, 0.05593529095086305, 0.0904234834337594, 0.15375429260045972, 0.25639808276676557, 0.3808767749090405, 0.4982031624147107, 0.5558911035613939, 0.5162233401100484, 0.40365404388242265, 0.2694671273577923, 0.1629172797181076, 0.1127961610307011, 0.11589480007822457, 0.1442268380545293, 0.1399078586648424, 0.08268661374718679, 0.016186878338586543, -0.0398537398212834, -0.08089834875902265, -0.10043836222853642, -0.10692751698638692, -0.11578251218975091, -0.14117773454223867, -0.2091062490916372, -0.30895595549340776, -0.37606178029261106, -0.3400899288754778, -0.1818088562427835, 0.006573575090122539, 0.09960856536940745, 0.06857661946972161, -0.07040889949768027, -0.3112439025280642, -0.5516923424709932, -0.6788224616290806, -0.6480632324383873, -0.47558737736841034, -0.19335789963220223, 0.17156236171401276, 0.5706170670505798, 0.9251227107982047, 1.155963876167707, 1.2229369697096901, 1.128155137274232, 0.9132724584260364, 0.6358227968037405, 0.3273167729035495, 0.03551096948200565, 0.06587813064072613, 0.09218868855087857, 0.13978784611527076, 0.2228664414378764, 0.3175380869255544, 0.3824203902647681, 0.36751786715523294, 0.2612324047521611, 0.11601257934651106, -0.017234670689559035, -0.107589013475744, -0.1316720444670926, -0.09448717791178443, -0.03409103999313449, -0.005487252949134179, -0.02584033337049584, -0.05955676599800484, -0.06470295772426829, -0.021912419526409953, 0.07322701125148517, 0.16848407460009737, 0.2321528142373828, 0.2510678642681981, 0.2046472343338126, 0.10952892086874666, 0.010536695282745078, -0.02856298545717535, 0.03785087326876806, 0.150021312043908, 0.19307987972907942, 0.13504569746988335, -0.007386029585349352, -0.24734245002269079, -0.49948286506489514, -0.655735731011502, -0.6680832643887695, -0.5328918228925054, -0.2787227842598344, 0.06811889426297826, 0.4635718400617264, 0.8263068310154903, 1.073896901202293, 1.1601697631462422, 1.079440094598318, 0.8722304001313901, 0.6027791812335325, 0.30831699773496973, 0.041992404802794196, 0.07423251951201798, 0.09450405040587471, 0.1293681583072327, 0.19663163127244224, 0.2687143105155018, 0.2963866207570798, 0.23269308934491387, 0.08233200263931591, -0.08490870031149303, -0.2163804450313613, -0.29114107734019173, -0.29251451462223044, -0.2348849260858088, -0.1609581878776534, -0.12107694125438123, -0.13195706838296728, -0.1503997539120378, -0.12776379051990888, -0.037951428670172704, 0.12933839503603103, 0.31851481360933015, 0.45326907375059944, 0.48560863579479524, 0.4520228249831801, 0.38147087694417436, 0.286492500769614, 0.21594393448667634, 0.21102061181564877, 0.24066946922569205, 0.22208522704749065, 0.13901943247776694, -0.0022228661160395833, -0.23793868040629207, -0.49620476974784117, -0.6708126661783927, -0.7173632893045544, -0.6130573816534959, -0.37895764949761557, -0.051700844459539194, 0.3251650650684749, 0.6773350653236259, 0.9289570668106846, 1.032438367231411, 0.9733070913980683, 0.7868032860652265, 0.5395016209386753, 0.27364464339582417, 0.04719496965373165, 0.08164858842210473, 0.09891669260462398, 0.1263464119486255, 0.18439259814950948, 0.24210222577386406, 0.24678133214290046, 0.15866779735650396, -0.010193126469617744, -0.18540719585433563, -0.3087682582118575, -0.36343272541821386, -0.342830984246302, -0.26902984054479273, -0.1930055092664936, -0.16634301423354192, -0.18853572949646036, -0.2011217646496688, -0.1657021875136212, -0.058317520485952706, 0.10770371144492183, 0.27218817719226646, 0.3343785921001235, 0.276391769475692, 0.20614412899864834, 0.19308561985656653, 0.21900495266638326, 0.2497545833252708, 0.28186433315059384, 0.2904947979092561, 0.23041346345105457, 0.12049935949147807, -0.032432270488568324, -0.27760125092971055, -0.5502560241720795, -0.7431385669062712, -0.8162531717103902, -0.7333895911134377, -0.5056295493422733, -0.18888150568555, 0.16469306280534318, 0.4943446485193663, 0.7388610360656738, 0.8542958798971246, 0.8199594423466156, 0.6636838888467693, 0.4506409743622566, 0.22608775021829022, 0.052441886851646004, 0.08988247705070415, 0.10779047212445934, 0.1345711208559796, 0.1909667111991921, 0.24206256835296577, 0.23448255272632937, 0.1374155358596945, -0.033634644891603886, -0.20686196385529973, -0.3205759822669476, -0.35572328639207806, -0.30766518370314566, -0.20851884564688405, -0.13290426182892653, -0.1332574230312099, -0.17551324057340842, -0.17656915962055708, -0.10264888903766856, 0.01748406315876834, 0.10989097528702191, 0.12377623941866078, 0.03187717251211999, -0.127850597562554, -0.2447571851419035, -0.23821606090277309, -0.12195463254819187, 0.05027933438651017, 0.23230631492690418, 0.3192082274707296, 0.25313653966985517, 0.10715493821722516, -0.08159187816079962, -0.35240513958549485, -0.6442474745017019, -0.8591750318871335, -0.959037254517099, -0.8938079240412423, -0.6630292051694635, -0.34340690570168286, -0.006259889091158033, 0.2984765684950462, 0.5286240591785754, 0.6488837035235473, 0.6369593686607024, 0.5148807067666262, 0.3442781152204559, 0.17001752754913427, 0.05880076322876324, 0.10026644951188715, 0.12126119437035132, 0.15235384236252814, 0.21448050534850832, 0.2672581605837699, 0.25487293334615146, 0.15622922417707086, -0.009469252115685584, -0.17676906732712108, -0.28095332892099867, -0.29766840512950704, -0.22697598824998147, -0.10983308804325286, -0.032940982258138456, -0.04731636609091323, -0.09090119291319564, -0.05113249344877766, 0.08951503673637416, 0.21021352553697614, 0.18628891686475446, 0.030477899218313176, -0.19477238216775622, -0.4342173702238085, -0.5945711898387553, -0.5948201247309192, -0.4455123169024672, -0.17923512271756709, 0.15755823522772608, 0.3707128568557275, 0.3131553472768813, 0.11278326269838822, -0.1378375883738552, -0.44711995544638944, -0.7590103374660975, -0.9962157507143865, -1.1240619895119501, -1.0780999495721635, -0.8436513414347488, -0.5125098016266723, -0.18019562910989495, 0.10490482424157724, 0.3176166474555647, 0.43547459364953406, 0.4403528983092685, 0.3521544436215002, 0.22833302189669644, 0.10972640447101596, 0.06659795460645722, 0.11277687475192896, 0.13777450193028565, 0.17441522410809468, 0.24501812605500214, 0.3059985791487504, 0.29578541366305844, 0.19899689882274163, 0.04213794670861276, -0.11314117571067904, -0.20573106673455194, -0.2133367104496243, -0.1388996430451289, -0.019998673350172594, 0.06083690096912379, 0.05535804565255314, 0.03731434597485046, 0.12955400497424105, 0.3260125580039974, 0.4358300206584845, 0.2810093170087474, 0.007619927154153513, -0.2879234201386722, -0.5590270532572376, -0.7368066794207392, -0.7505483580573633, -0.6080014581997248, -0.30741196365818685, 0.1274544471147512, 0.4468991449924934, 0.38340497008181545, 0.11369896789138541, -0.20658531923258527, -0.5511992020381505, -0.880439647319719, -1.1388917359373196, -1.2909019939068, -1.2633977464917119, -1.0303857657286195, -0.6881241586980968, -0.3528307086385029, -0.08076071124792544, 0.11419854896148926, 0.2243872859656849, 0.2408534585280302, 0.18499522500556512, 0.10968933570316247, 0.048756323585549266, 0.07511769531459057, 0.1265461539890226, 0.15544297805292473, 0.1949772090645591, 0.2698808411613319, 0.339277779185191, 0.3361236527014318, 0.24495204737563955, 0.10021954116095622, -0.036137427190947743, -0.10884580262039127, -0.10527833205758266, -0.04035681591270363, 0.05447669472212901, 0.12138260569265703, 0.12403274056724421, 0.12224906968311137, 0.21385461459325228, 0.40599887250032557, 0.536245530183694, 0.34713785457300095, 0.025852501973505467, -0.26496046146139324, -0.5101797768768801, -0.6860507735462241, -0.7238132279580861, -0.6058491819671186, -0.30936545607279614, 0.14181798028134654, 0.4766253443466632, 0.39065365304428357, 0.06997992930473257, -0.2976717804202231, -0.663473596985851, -1.005626414031433, -1.2791548301712528, -1.4433193398637767, -1.4280866873303333, -1.203024050254165, -0.8585502054234311, -0.5216758706962686, -0.25992729485738364, -0.08199475943601274, 0.018644808226535137, 0.044317907800083774, 0.019960943598211482, -0.006259063492721137, -0.009925984538049359, 0.08163056500593249, 0.13791354250389792, 0.17125858483140752, 0.21006389512401868, 0.2790765357434749, 0.3479931024123161, 0.35110799248281077, 0.268517284210037, 0.1401458894056071, 0.03172394853152473, -0.009389830157293404, 0.014906396352863263, 0.074668540461133, 0.13513313910564928, 0.1567174910672268, 0.11806283080711516, 0.07682067320331354, 0.12585877650029662, 0.2789343444112088, 0.42953456801245166, 0.3331797016256399, 0.053391341893634724, -0.17986594996164273, -0.3536829816746768, -0.5039824652350626, -0.5690075140157359, -0.48987182750268443, -0.22740153794613732, 0.16861862088651178, 0.4300173427070765, 0.31074165349269345, -0.0321375271088319, -0.4163230890086957, -0.784682313247973, -1.1279392011227978, -1.4034534897437037, -1.5658373848316525, -1.5558051194238016, -1.345047740517669, -1.0117377310764581, -0.6824655785000069, -0.4334205295728373, -0.2717726168457489, -0.1798446522248474, -0.144664570698002, -0.1372091862527136, -0.11479963930691524, -0.06390935642072458, 0.08290134681409288, 0.1410552265741853, 0.1785327673310351, 0.21472813289247816, 0.26868580640673273, 0.3236853806194411, 0.32541058512652266, 0.25160268403069846, 0.14142980455222331, 0.06394149144458429, 0.06433175070886064, 0.12643064152034555, 0.1996432547567701, 0.2340399262278431, 0.1954441008309022, 0.08264773108915205, -0.04509030344614165, -0.09078998269050469, 0.0006250718289233306, 0.1441235082880671, 0.15539302107838895, 0.007438055671153129, -0.11152583414435609, -0.18197476322489928, -0.28334959157758693, -0.3580126738321705, -0.3152884513127318, -0.10954681325554096, 0.18905656609955562, 0.3416816643047139, 0.18009368753447466, -0.16355690343905996, -0.5335535645773836, -0.8886157758093842, -1.2255726580928932, -1.4907716393253356, -1.6399931244886399, -1.632516551558566, -1.4428863126098777, -1.133732824489975, -0.8239153818377447, -0.5929269829965428, -0.4476513039831974, -0.3629309772352495, -0.31718143053336845, -0.2785680303904941, -0.21102505011455977, -0.1112921586633173, 0.07770972339946022, 0.13235088376912715, 0.1711084019929174, 0.20392945022223657, 0.23896099215312167, 0.27136058376641653, 0.2642840351196888, 0.1970249352889946, 0.10420073734324889, 0.05327487307965857, 0.0909614060794537, 0.19662527022591442, 0.3046115637412186, 0.34354246163788676, 0.2571386349886559, 0.05746727965076946, -0.16386862080424197, -0.310846615523487, -0.31501877713461396, -0.20003555299005896, -0.11560393627912764, -0.11738779217163922, -0.1045888874429312, -0.06458063065929455, -0.08725430907924744, -0.13618664215511642, -0.11741479501844189, 0.022328615445959894, 0.2103012973282704, 0.24913480766393853, 0.05021039890361422, -0.2693349053478416, -0.6050525619765562, -0.943822056704064, -1.2685630997394075, -1.5150473945856553, -1.6522573349980554, -1.6518254236949181, -1.487374146249584, -1.210304516249601, -0.9298540367402877, -0.723594296956862, -0.5967484606102726, -0.5188635361739323, -0.46267622504741024, -0.39625527186534587, -0.2908163110179796, -0.15090230874481977, 0.06849196066047683, 0.11486709535000436, 0.1495638114334389, 0.17743624238564845, 0.19528041581711011, 0.20411330023214236, 0.18540155522142773, 0.12422515308337877, 0.047713420575285936, 0.013980979596705688, 0.07085712124152221, 0.20590752807117266, 0.348101680648872, 0.40907347019890766, 0.3098375562902555, 0.05183660505006461, -0.2512746923593735, -0.4765320601019498, -0.5653695873685305, -0.5047987131125444, -0.3862951661033756, -0.283182560875414, -0.15456361069780258, -0.016651865273653508, 0.04625797203424559, 0.04572234470573704, 0.05723317681810703, 0.13968132326754462, 0.22999900223307299, 0.175865632693111, -0.038921311473459105, -0.320229711008896, -0.6159614163567627, -0.9333438791721197, -1.2394994957240182, -1.4708745267027283, -1.605304722666507, -1.6148978394510118, -1.4743386283560511, -1.232335949236791, -0.9886688154210138, -0.8132781276381588, -0.7076298495495843, -0.6375055911569846, -0.5727004414838648, -0.48407936158070186, -0.3504432271077182, -0.18112623037614087, 0.058670527642495134, 0.09588644121969764, 0.12280266272617361, 0.1429347397398517, 0.1464518227713246, 0.135741982345173, 0.10617428830406736, 0.049982008719103174, -0.010900150612291047, -0.03164894657579129, 0.023816965737936447, 0.15047090685921188, 0.2929770447137661, 0.36575930794961814, 0.2779375715210591, 0.017793124642761057, -0.31023360845490455, -0.5798427374157802, -0.7233750397465075, -0.7230310174035948, -0.6099265812479624, -0.4451245288874921, -0.23383244979762113, -0.024788993581823134, 0.10297736415297007, 0.13924662640751653, 0.1450658111361103, 0.18872704405746665, 0.21964419846910482, 0.12539052763871275, -0.07591934771408397, -0.3118372850685573, -0.5676306117106868, -0.8619411206895713, -1.1490993520260424, -1.3704016633287652, -1.5067396811337346, -1.5250656308215191, -1.4057762575757122, -1.2000120830572187, -0.9964968120701498, -0.8549700221752533, -0.7722161108047231, -0.710923045845714, -0.6403645183246328, -0.5364286443271009, -0.38537894010406293, -0.19905059415134632, 0.049238578261609926, 0.08062198716018606, 0.10104522827765954, 0.11255212454530006, 0.10484620463389308, 0.07911598992410113, 0.03877027112131653, -0.015873074969367983, -0.06707180408342527, -0.08050989571910594, -0.032962534124140075, 0.062393924922049294, 0.15956872872850306, 0.20410829388522864, 0.13104378147721638, -0.09122160253140768, -0.38719456218152254, -0.6504068579549576, -0.8107338623528343, -0.8404794948037003, -0.7421791185895633, -0.5453133704937383, -0.28782944808024974, -0.043621624623803104, 0.10637937835076841, 0.14706823699628818, 0.14514721319314436, 0.1711612286987778, 0.18258849342070307, 0.09882002301749235, -0.05953647245360175, -0.24371341858243495, -0.4681778183987365, -0.742443424788957, -1.0107874156196939, -1.2259887225161619, -1.3654293393725112, -1.3895421190717865, -1.2896675261567212, -1.1193196821253095, -0.9557505085040691, -0.8484987811088894, -0.7881135268156886, -0.7353558078970935, -0.6619802070885392, -0.5501617050736167, -0.3928367556241221, -0.20272301396764222, 0.040280350348339246, 0.06977838805251996, 0.08856849056666131, 0.09444038907398822, 0.07998430397961635, 0.04627454297138646, -0.003205695221368245, -0.06252358354581873, -0.11260316860000887, -0.1289978928976431, -0.09833215326972812, -0.03621049758751503, 0.011334079969325209, 0.003950187591798534, -0.07628966753335678, -0.25222019831131026, -0.49384961599342275, -0.7161537352479521, -0.8564022855059811, -0.8778376552539793, -0.7758071087790035, -0.5656690981253252, -0.2987912864644444, -0.06296824692188342, 0.07160127691988642, 0.10468021519092989, 0.10648234748298882, 0.13342122947940355, 0.14852650257223607, 0.10095601765196172, 0.003083830351778402, -0.13221513754473863, -0.3335904600395215, -0.5892305136448011, -0.8435493809902606, -1.056322076957357, -1.1945975885352849, -1.219609895513743, -1.1368807151643734, -1.0001615713353043, -0.8756173543945157, -0.8014345420594631, -0.7609036472338631, -0.7153255893711098, -0.6419082535059405, -0.5293577095651176, -0.37553138505869815, -0.19328580619509558, 0.033095852356068274, 0.06263791715414674, 0.0831928674114226, 0.08865851912365044, 0.0739903411144144, 0.040614579971259696, -0.010554461614114326, -0.07593881722433211, -0.13465228153767403, -0.16253352968270188, -0.15415045146973289, -0.12746118658397634, -0.1184710356864675, -0.16316929883768508, -0.26248209114100873, -0.4076534408882979, -0.593928097614642, -0.7696935347361789, -0.8764442867522947, -0.8794230711696941, -0.7715201648371713, -0.5715146099770991, -0.3262250855899291, -0.11970565870291426, -0.0007356635549997703, 0.04298914830011427, 0.06599414587504152, 0.10198220560800371, 0.12705740819215042, 0.1182064333806192, 0.07529819775093405, -0.022639286411971115, -0.20335069325138527, -0.44100257876839893, -0.6820110013783593, -0.8844037199390544, -1.0092139277506895, -1.0281484168298036, -0.9603603421377662, -0.8563570463220412, -0.7703546714689858, -0.7272468810619198, -0.7030328611726127, -0.6632423440947556, -0.59271284423194, -0.4852475440192259, -0.34184343552699115, -0.1752890340323359, 0.029214481275761466, 0.06024431636405474, 0.08396820999738464, 0.09190395588979616, 0.08187330518253524, 0.05620420144746023, 0.013375604607900205, -0.04993738901343476, -0.11941464338839618, -0.169124448761551, -0.18733583055557967, -0.19153316839457007, -0.21360129095294136, -0.2789189746419879, -0.38679383777217546, -0.5137849022297567, -0.6520150067087175, -0.7847302609116094, -0.866943800700909, -0.8658643113844052, -0.772933935744424, -0.6060830673930145, -0.39973090002284684, -0.21627904507073184, -0.0950345586690679, -0.027025663856272645, 0.01784284870912073, 0.05832534421372693, 0.0913849220411546, 0.11112636485926083, 0.1028744748571328, 0.02755141945537143, -0.13184582576919512, -0.34327588185023816, -0.5552105567156438, -0.7262789148638384, -0.82115495239364, -0.828223258456467, -0.7757584464615849, -0.7054139698623898, -0.657258529368551, -0.6412589815045202, -0.6288860803503143, -0.5947047702657914, -0.530628802657011, -0.4321773582587816, -0.30250154956114267, -0.1545480184601683, 0.030850373778213116, 0.064258432211574, 0.09161757692005307, 0.10502000294380971, 0.10149662844009856, 0.08366011941375828, 0.05336651961068701, 0.002906011910742175, -0.06627359046799094, -0.13685406134617586, -0.18967235184359788, -0.22609182848730652, -0.27202620214126016, -0.34941152218261173, -0.4553190968155112, -0.5649519373734126, -0.6628592039066412, -0.7507556814941927, -0.8113365611620718, -0.8160826170196637, -0.7532939208200714, -0.6333862743950699, -0.4779291041076402, -0.323501639548168, -0.20596168707625997, -0.12603341457314693, -0.07080596050593417, -0.02671160429069745, 0.015704639452975185, 0.051550156262420876, 0.05440591733563669, -0.010354303108984067, -0.14257919007817518, -0.3099864896832678, -0.46837640528594304, -0.5855222815723853, -0.6390198940024019, -0.634830688595516, -0.6005635804407737, -0.5641691138783437, -0.5504544157498018, -0.5551822968486242, -0.5511157137124453, -0.5239115101979399, -0.468769677412868, -0.3808068480202154, -0.26542308147771837, -0.13545504004076822, 0.038820986781094954, 0.07635357273988327, 0.10706900093457569, 0.12752632811863174, 0.13343873773681217, 0.12204882970793848, 0.09797423405336247, 0.05879294671863488, -0.0010238478800956802, -0.07780796532143608, -0.15824024014032376, -0.22894719412066705, -0.29779779158765673, -0.3820517945993249, -0.4819624467321209, -0.57491116776631, -0.6414129655152295, -0.6861386695228173, -0.716337729133435, -0.7196716614132705, -0.6830095862057346, -0.6105304206697002, -0.5142563757770914, -0.41075884950763014, -0.32273722988168785, -0.2532602978660276, -0.19748656150811658, -0.14701137142426204, -0.09786251830922506, -0.06147972941652767, -0.06306098006465796, -0.1167694921657257, -0.21116241756196824, -0.31750877172000924, -0.40554696958301156, -0.45940273904821105, -0.47350905098617707, -0.4642781095315603, -0.4495911713379039, -0.444309013165753, -0.45859634443135805, -0.4773205073746093, -0.479530780069923, -0.45948990040353693, -0.4125877047780054, -0.3347373856105621, -0.23329760638253713, -0.1194838102365833, 0.05138368237169502, 0.09601164763829334, 0.13127469923724178, 0.1579158775618926, 0.1731889800385708, 0.16976557861739783, 0.14720929961413548, 0.10766536194697414, 0.04960426204722421, -0.02661767428679665, -0.1176645855840257, -0.20994600028033053, -0.2960163081162886, -0.382319675313763, -0.47183248941797856, -0.549715534529978, -0.5953183358217916, -0.609424577109819, -0.608579526496016, -0.5997560866139515, -0.5759498359871249, -0.5382748698867706, -0.49377354262231044, -0.446946885666283, -0.40346884500614105, -0.3613992816899883, -0.31748507651336766, -0.27131517467826627, -0.22928205838690469, -0.20395897258016654, -0.2083693922053309, -0.24020469824832033, -0.28710172725316246, -0.32684932125482746, -0.3475417025554495, -0.34833396399274563, -0.3371103837014663, -0.3299851600750507, -0.3333096317031727, -0.3526901758566905, -0.38535004086711355, -0.41160106951410197, -0.4182938264344263, -0.40336145908877913, -0.3620921731277632, -0.2934443723841878, -0.20539584752040438, -0.10591581646725286, 0.064766563576385, 0.12005719865706638, 0.16262063007207705, 0.19442720556847506, 0.21583505282074317, 0.21936019338661586, 0.19775003156786322, 0.15112308808778435, 0.08220716689214251, -0.0019458999858075908, -0.09659963034122777, -0.1944284566203237, -0.2834962909630278, -0.3624971472658879, -0.43427711696143684, -0.493657850403682, -0.5242348295569669, -0.5216595914904244, -0.5010262876053766, -0.47845550467541864, -0.45773115041632295, -0.43899216265694635, -0.4272426151587561, -0.42111639190789496, -0.41684565215373715, -0.4083897060678377, -0.39056084975787037, -0.3665367131458919, -0.3435119297270364, -0.32965043556321405, -0.3263490927793552, -0.32861109884816014, -0.32794852899717064, -0.31356470268715, -0.288095489071483, -0.25903856203231823, -0.23995036286792137, -0.23891888350047175, -0.25531057268240737, -0.28892833630377573, -0.3287926623630658, -0.3572647127213613, -0.36658077716673987, -0.35403550077625184, -0.3162321022373003, -0.2560391630060289, -0.18043151355716933, -0.09369516768220912, 0.07450895415645796, 0.14255513279780901, 0.19585313870975204, 0.2317966571746086, 0.25433378416811253, 0.2609815497189703, 0.2401023626658239, 0.186437922454882, 0.10404615568511129, 0.004083917971216007, -0.09899760330307454, -0.1965231993662297, -0.27884054364870753, -0.34151047736971546, -0.3887308673508581, -0.42315617570605, -0.4375346256202452, -0.42448457852276866, -0.3934480434734642, -0.360616730536963, -0.33752675091871565, -0.3267540252138816, -0.33142827214417186, -0.34883080707202535, -0.37196356757502513, -0.39365540092120965, -0.4070452794233477, -0.41112937677696226, -0.4074975019996861, -0.39876317899190666, -0.3824091282659149, -0.358080910003035, -0.3232359747127759, -0.279274259016801, -0.23405559732114417, -0.19798228751428743, -0.18149561461409597, -0.18529180215638164, -0.20769594754013374, -0.2448519547696376, -0.2836127636998507, -0.3108722544776219, -0.32024713622972645, -0.3076493536236677, -0.27282415855281017, -0.2210841176581728, -0.156654914703524, -0.08151633373633704, 0.07850351906322928, 0.15598977215052048, 0.22084431997712248, 0.26202047670880646, 0.2811247731644126, 0.28297107316738973, 0.26038078075082577, 0.20342921331359254, 0.11428342712956005, 0.004196198611672372, -0.10865367436554528, -0.2074900997642176, -0.28283878672710955, -0.3297881099388384, -0.35295847042576445, -0.36132753940438683, -0.3571108887269877, -0.3349570183685936, -0.2968755421747487, -0.25562092691345856, -0.2250385580825078, -0.2136103315807743, -0.22348822429066514, -0.25326202461000524, -0.2941278808508356, -0.33776643559273334, -0.3753016074576386, -0.3999392227819118, -0.40857358672233407, -0.40024645424986693, -0.3749368874857172, -0.33594985501072355, -0.28750284970656115, -0.2368164019554374, -0.191392828535068, -0.16048103209730513, -0.14849061776931632, -0.15440918807755138, -0.17703822887268658, -0.21035591074661114, -0.24345425990483924, -0.26651751462082696, -0.2727313190914576, -0.25899589288893277, -0.2282539588837565, -0.1853077866274091, -0.13143824530531617, -0.06813180065717561, 0.07703561754551576, 0.15545286030907582, 0.22629877224225878, 0.27420530107937935, 0.2913319562172257, 0.2820150034239554, 0.24999278686020626, 0.19080079114166099, 0.10354646381338493, -0.002804470110122973, -0.11361205470722287, -0.21036330632555233, -0.28001688092792, -0.31784804288875185, -0.3262233907196308, -0.31608322868578703, -0.29635615576810126, -0.2668428630092342, -0.22510339783160316, -0.17772248807657165, -0.1375641890463191, -0.11732333699561222, -0.12250137545676436, -0.1533391810019313, -0.20136886664316644, -0.2556117812744307, -0.30508791131607266, -0.3401244614843103, -0.3551942327238728, -0.3490534064251715, -0.3240059333104831, -0.2848292376833005, -0.23932690538802853, -0.19490202867367723, -0.15830640257145098, -0.13547256835703164, -0.12772128238475755, -0.13436884569861088, -0.15303992628695395, -0.1784809700138051, -0.20358824446132948, -0.2200559675767314, -0.22144955921501816, -0.20734434280112848, -0.1817461655899797, -0.14740034095853077, -0.10422629240239265, -0.05373867509442691, 0.07089402261613957, 0.14157174389937177, 0.20876520379343746, 0.25977193342495375, 0.2787413712360179, 0.26193324694185893, 0.21737758632103216, 0.15224903489941244, 0.06987903260242202, -0.023567036158546995, -0.11819751370673391, -0.20159945916933517, -0.26132471733722396, -0.2919384270509739, -0.2943862947232166, -0.27643359814552837, -0.24899058561750415, -0.2158545360948235, -0.1751659286290446, -0.12745512774604015, -0.0819564939179513, -0.051098543024677555, -0.04541253032804374, -0.06731864347079226, -0.11104396174705071, -0.1639878269087602, -0.21485804761747598, -0.25323879020766876, -0.2730301256761085, -0.2732103874081579, -0.2558934025587664, -0.22601989898624664, -0.191013750580203, -0.1576496682120508, -0.13170690645142488, -0.11634845776264247, -0.11204884745635214, -0.1174057607445315, -0.12979861605795284, -0.14669479812226294, -0.16346586950593853, -0.17297892847061022, -0.17059523094951148, -0.15738803565177442, -0.13669430817690328, -0.11007527915384382, -0.07741628954584373, -0.03975944940705488, 0.06042843130131666, 0.11857387200520642, 0.17410013638820157, 0.21985128122249872, 0.239860963668548, 0.22379835416232827, 0.1753840853242464, 0.1061997792251901, 0.027665404882893194, -0.05231852380083605, -0.12663488666575215, -0.1885293112072007, -0.23176100875726294, -0.25183034273730764, -0.2500495507309823, -0.2314583642459778, -0.2042038136621211, -0.1729884805382933, -0.1375152961080159, -0.09631859584574616, -0.05336075438025988, -0.018282993260063846, -0.002774060136382851, -0.012546115043521754, -0.04425471165235573, -0.08729697594994452, -0.13162559778523772, -0.16797477134991068, -0.19030100887034848, -0.19674431534555367, -0.18883345583911046, -0.17048545965159784, -0.1475302202464693, -0.12576901836476248, -0.10900043922934466, -0.09892786990718411, -0.09551249956685243, -0.09769071624879548, -0.10454162983215724, -0.11481344921038726, -0.12465614080171691, -0.12873299975616714, -0.12433046794581129, -0.11256532797128117, -0.09593677991932101, -0.07603301460882106, -0.05294945943456485, -0.027051357118921746, 0.04625150519305883, 0.09063171145524096, 0.13128124893382176, 0.16457247358211574, 0.18125483106293755, 0.1704609317380503, 0.12953424783475076, 0.06629601280150592, -0.005189766968925995, -0.07276188397667721, -0.129345625027055, -0.17055152679194677, -0.19530250541450767, -0.2030993419205428, -0.1962610779167483, -0.17978670673335545, -0.15814659832876996, -0.13418535124784925, -0.10753956218314395, -0.07710873782533596, -0.04325862235601528, -0.0121316810808431, 0.007057463395441495, 0.006952759313590075, -0.01093541218243381, -0.04002323684933148, -0.07261704070295245, -0.10187646196538501, -0.12200650471694652, -0.13069838409524287, -0.12907897429841023, -0.11991595230098735, -0.10744092802444805, -0.09534701587904333, -0.08559539792048063, -0.07875492768926143, -0.07474100630052431, -0.07421337440897115, -0.07771731707841045, -0.08367349323062993, -0.08875816988276174, -0.08933413990354355, -0.08402640602232941, -0.07398606567188797, -0.061202730229069184, -0.04724271696605151, -0.03230449007048615, -0.016313522722831934, 0.03049434507081566, 0.060567533070431626, 0.08660685703935202, 0.10602696876033431, 0.11655432427770578, 0.11109029393966963, 0.0833016125798188, 0.03605963900803672, -0.01955094093497533, -0.07104217935039696, -0.11082980341081378, -0.1358458180710796, -0.1465218784165457, -0.14559480582885534, -0.136312293798974, -0.12350482056567234, -0.10980450689696955, -0.09538490016443545, -0.07930577789356553, -0.0605446283909103, -0.0386610193781604, -0.016455853183220525, -0.0001025686468282567, 0.004603160758360497, -0.002550930889295282, -0.01812821188288045, -0.03771496346865249, -0.05678753214812194, -0.07066975703703339, -0.07746807897676694, -0.07789150435502183, -0.07406958843858846, -0.06847321882309601, -0.06293554347412557, -0.05838709961084983, -0.05440833190084417, -0.05091524370706795, -0.04948371329491625, -0.051068455313373805, -0.05433242260821361, -0.056611723875179744, -0.05544050459864047, -0.05043230913336198, -0.04293651498763213, -0.03438225065539407, -0.025740731465535426, -0.017209378018468845, -0.008571104180327787, 0.015034605209912249, 0.030186605880999726, 0.04278681439795374, 0.05118150834558586, 0.05585779467416651, 0.05386617084055534, 0.040120450595223386, 0.01503050878609881, -0.015478473445356473, -0.04384432845002684, -0.06473437537107908, -0.07665246380160458, -0.07990506521148899, -0.07692742881326652, -0.07041231457364489, -0.06328695054006497, -0.057071686071054624, -0.050930845809075816, -0.04408074987925356, -0.03557852175495891, -0.025282796903013875, -0.013961799912264065, -0.004815989456940589, -0.0006145075641617087, -0.0022538133436884773, -0.008188581425561546, -0.01680236916575495, -0.0256844620501862, -0.03228273264625289, -0.035504861153115054, -0.0357600172611272, -0.03434923898694672, -0.03227281318254339, -0.030379392761774905, -0.029027746677740617, -0.027558242351438954, -0.025773357531479864, -0.024773121374151556, -0.025263575325308953, -0.02672126298708498, -0.027532305750313738, -0.02634485704145585, -0.023254022372237133, -0.019236535969072092, -0.015077300176749938, -0.011069174717436235, -0.007292746500155012, -0.0036079757490100656, 5.4851202254792035, -0.010615819984669157, -0.019928273262239646, -0.026909476567115644, -0.030760935235294332, -0.03128305209800454, -0.02870083031876619, -0.023171435089892705, -0.015391494298439294, -0.006185914089346551, 0.004895285516143444, 0.019190992515667894, 0.03699180824953424, 0.05738761513763937, 0.07928544266784898, 0.1003870383988707, 0.11711317115555989, 0.126105433660789, 0.125256246235757, 0.11452916904090542, 0.0954959786151672, 0.07026000747937208, 0.0395345440119671, 0.0032702669668071413, -0.03802041904102115, -0.082374407764183, -0.12634568382523903, -0.16636288188956483, -0.20023905823068203, -0.22752968865831974, -0.24866384125671173, -0.2649481751386248, -0.2777316483767485, -0.28573512323576994, -0.28504847955289636, -0.27361426316923715, -0.2515979250343533, -0.21920955470765358, -0.17656768435448675, -0.12604233102338483, -0.07400587408503413, -0.028585474897674826, 0.005227606754107228, 0.02758992501660755, 0.04064133272178401, 0.04537031068596887, 0.042091267272438435, 0.03191274369887864, 0.017022036052498933, -0.022214930563169193, -0.04218260774815857, -0.057940827904019006, -0.06755375252592931, -0.06972204006085542, -0.06436646580909122, -0.0523970060512897, -0.03548441646922484, -0.015312358036617601, 0.008648312744430252, 0.03851284126036969, 0.07449487314850509, 0.1148380345472791, 0.1571208186230117, 0.19664029039180816, 0.2259830907408012, 0.23850508763775866, 0.23140238561779888, 0.20711855655059785, 0.1712546427994465, 0.12826692207689014, 0.07738621013009522, 0.014349066464400359, -0.0627861220201602, -0.1504822355308787, -0.24089030328379338, -0.3255206630395822, -0.39876574453801394, -0.4587122035440809, -0.5048682820964471, -0.5382357879497176, -0.5610354483871469, -0.5724320160732796, -0.5673880779849094, -0.5419720462264199, -0.49578813833394847, -0.4297292710074709, -0.3456146587348594, -0.248700203471324, -0.149819690542476, -0.06263346993906918, 0.0036922282091194788, 0.04820916008614449, 0.0741290140916542, 0.08390078104653949, 0.07864676299952121, 0.06024229732903089, 0.03243675507683257, -0.0348526526951253, -0.067306169375141, -0.09447710680183595, -0.11244677020028482, -0.11764395906746243, -0.10921032447209746, -0.08923802282783815, -0.060947985148101645, -0.027274075412629786, 0.012003852892917305, 0.05921456750008271, 0.11404480534000458, 0.17361728232733142, 0.23419808659390914, 0.2883102824093729, 0.32368252556792254, 0.3303302643831049, 0.30729112919099955, 0.26374137885734994, 0.21312013226206372, 0.16339866579331094, 0.10977664591062798, 0.03797637569288038, -0.06181460814293468, -0.18691743645650727, -0.32454632837328473, -0.4591655221430267, -0.5792388660948188, -0.6793464598040331, -0.7569120373480962, -0.8109660302303784, -0.8430348952995579, -0.8538975989758654, -0.8403176521711674, -0.797928546374232, -0.725416238455241, -0.6251502671927354, -0.5028325792731442, -0.3672110187684127, -0.23062861801839438, -0.10815307122726575, -0.011649602572230171, 0.05513740853235976, 0.09452872086220127, 0.11048134396287673, 0.10580842376767023, 0.08266179510658193, 0.045154811995183154, -0.047812418063009565, -0.09373249725192828, -0.13374850410789665, -0.16111083838722223, -0.16947179855765612, -0.15717560586159543, -0.12759792074458393, -0.08611383976439727, -0.036999582512912466, 0.01977656460092177, 0.08594223843647851, 0.15962049935854103, 0.23705897471082626, 0.31415703909116116, 0.3796737052043088, 0.41507655464999443, 0.406903613008823, 0.3577498803335628, 0.28665290156406137, 0.21886119421958736, 0.16896026675512868, 0.12840626931473006, 0.0695778135961641, -0.031401864267802095, -0.1781293329885944, -0.35524935820876796, -0.5394280828266975, -0.7104716556538786, -0.8568400599385989, -0.9730523088276436, -1.0555219086448695, -1.102720114181941, -1.1156706271800014, -1.0937443730279575, -1.0342195330634039, -0.9360731498880035, -0.8041033859902469, -0.6489134832510879, -0.48336778880446907, -0.3194330879348416, -0.16966127373497228, -0.04661049704258039, 0.04248301461972258, 0.09738227619696181, 0.12217188510396215, 0.1212954010324634, 0.09739451923547268, 0.05423255389989477, -0.06023433811031251, -0.11866334365544942, -0.16984872749377009, -0.20431229363994077, -0.21352015292854212, -0.19572097503823466, -0.15587048721491725, -0.10105294209694585, -0.036130938713530354, 0.0386976446406449, 0.12336089944168586, 0.2141591567792765, 0.308403479786398, 0.40199642445755346, 0.4785486942144629, 0.5126811518466466, 0.4864387647683196, 0.4043559580285502, 0.29487391863542556, 0.1985825553116184, 0.14250739827125128, 0.11942124860253597, 0.09058141418440471, 0.014417758764001334, -0.12687626546420053, -0.32252503286127215, -0.5439107130039634, -0.7616160745037364, -0.9563032815871894, -1.1180362734169667, -1.239808937527272, -1.3149977648443507, -1.3411765631667483, -1.3186379179272816, -1.2473539918630894, -1.1287561397141632, -0.9708605492131615, -0.7888782732103744, -0.5999337066955381, -0.41639695773431334, -0.24721058083263228, -0.10299806395785799, 0.007208165178572009, 0.08024828107480687, 0.11786850526218628, 0.12421351152423205, 0.10301028530262771, 0.05854035223124336, -0.07157206551102488, -0.13941187137944042, -0.19650605020745315, -0.23218033414264955, -0.23763950409365509, -0.21240177625790968, -0.16298496034388385, -0.09706539171392958, -0.01918757556157252, 0.07029938091820527, 0.16961686697362682, 0.2754623609818812, 0.3876487972993103, 0.500448414474183, 0.5923586882248825, 0.6313356144830028, 0.5920982150449855, 0.4760985579967929, 0.3176388275562343, 0.17460991495753636, 0.09329821533178603, 0.07605634357349085, 0.08028721163711518, 0.04904173611692326, -0.05547987687404445, -0.2372543135772968, -0.4703086952934908, -0.719999148846869, -0.9594034995899373, -1.1718676550484697, -1.3441838179855417, -1.4624243730402717, -1.517255971969747, -1.5075698539956435, -1.435614115527081, -1.3062763239340147, -1.1309243201993155, -0.9285284104646917, -0.7199000647797837, -0.5201374815673682, -0.3370296143050837, -0.17746154278601706, -0.04929329261966967, 0.04259853367176433, 0.09660594661208477, 0.11401777054636347, 0.09880740267014898, 0.05723066787986223, -0.0808938355207471, -0.15300028239452718, -0.20816322365249004, -0.23728915272094322, -0.2341000798992422, -0.20045520908474462, -0.14404295277887494, -0.07254703242354066, 0.011508073005980625, 0.10906795688981692, 0.2182708428351259, 0.33815916550923997, 0.469098264008115, 0.6029279419656782, 0.7159938452929154, 0.770294262899344, 0.7299748133537489, 0.5872965461776608, 0.37777116655675913, 0.17456816812006098, 0.04617898844682743, 0.011660214346692744, 0.03456087246185639, 0.05166439681887334, 0.0064619633581285306, -0.12759286722309737, -0.34030345537806894, -0.600383139180371, -0.875453909855924, -1.1397904428949728, -1.3699266345729408, -1.541780825499937, -1.6375373721307924, -1.6532796533887741, -1.5931970601770808, -1.4653114060102015, -1.283662745006427, -1.0694234414278092, -0.8458228765743019, -0.6314474290755177, -0.43641503328232967, -0.2652732188933128, -0.12328996006456935, -0.015264315375893525, 0.05577987737206913, 0.08802818887779838, 0.08348180822626741, 0.04984523003096947, -0.08523294020574133, -0.1547952629324762, -0.20004726402492673, -0.21630023387965938, -0.20193407952092532, -0.16130944367872577, -0.1033310062241979, -0.03430556530936756, 0.04838189105543643, 0.14841882486164515, 0.26452145975069125, 0.3955386682710607, 0.5392096259124816, 0.6878658934163082, 0.8208450002961849, 0.8969858836825079, 0.869043121985837, 0.7155376458378367, 0.466626040554981, 0.20373105807152422, 0.015459165798885628, -0.05894558144968165, -0.040217754254410966, 0.013878548428958061, 0.03532305105455666, -0.02798243700574173, -0.19406629274028708, -0.4442302302835887, -0.742416056365689, -1.052152187910391, -1.3379268582835513, -1.5640418845983772, -1.704377859039727, -1.7515002030317233, -1.7116163784195062, -1.5957134510882924, -1.4197132768099545, -1.2053340891993227, -0.97621151776203, -0.751913505673991, -0.5458974422682636, -0.36452288340520794, -0.21191499731493835, -0.09197679626961007, -0.006436059975115702, 0.04273809892371532, 0.05466645617875523, 0.035760436237211576, -0.08088258020232772, -0.14020819158994152, -0.1697950264415977, -0.17053332599735907, -0.14575264335884391, -0.1027748493467515, -0.05149300020823825, 0.0071941181191214915, 0.08385429955245453, 0.18481571269821404, 0.3064969605944549, 0.44181808834187225, 0.5845421741763474, 0.7308569679007291, 0.8696080648128603, 0.9631030955004822, 0.9565295480742741, 0.8136716932823476, 0.5507591590872419, 0.24637739956823215, 0.0018380668728279594, -0.12365342052789534, -0.12950414551393408, -0.05899204760348863, 0.01801732795350803, 0.026960722127341515, -0.0818702493370201, -0.30899221180592623, -0.6161889534371244, -0.9559565007100165, -1.2822858983111296, -1.5500749958928277, -1.7270743840103995, -1.8032296759249928, -1.7857406962846814, -1.688050850005599, -1.5272037015305069, -1.3243008016135474, -1.1018399156761567, -0.877561746451093, -0.66527721574636, -0.4752382216375172, -0.3136551441575122, -0.18433326072084877, -0.08715506409313566, -0.021246255587601598, 0.011662549791275387, 0.01458080755390123, -0.0658594167474067, -0.1076877769923924, -0.11872563167586103, -0.10432244406121687, -0.0727218333029789, -0.035233448580140894, 0.00039669098770521135, 0.04358387662448893, 0.1140787218294903, 0.21910987086844058, 0.3471005163296723, 0.48015695034844336, 0.6060921507006958, 0.7259046695077691, 0.8437925723125229, 0.9362908835478433, 0.9522384001961787, 0.843244516426815, 0.6032907708511971, 0.2929583102459371, 0.012824306926857364, -0.16123576917498628, -0.2068324106550282, -0.1474612781221193, -0.04584096311750106, 0.01109292918482177, -0.04989111423127593, -0.2493628961611407, -0.5517088688380039, -0.9005357129591315, -1.2419612161788935, -1.5265543196221938, -1.720303448960127, -1.8141562907385445, -1.8159487250081092, -1.7391598165017232, -1.5997445538130413, -1.4167456505425966, -1.2109586999555357, -0.9981237567774147, -0.7891126762642549, -0.5954286808440258, -0.4265331344658432, -0.2878837724120293, -0.1796112740526306, -0.09797677408427424, -0.04246980388528156, -0.012783904618226642, -0.04069453294490219, -0.05948341588202762, -0.05124672509524885, -0.024124561770919983, 0.008171832384015289, 0.03126357825887447, 0.04443580798628265, 0.07099453583522829, 0.14045581809626073, 0.2575179526379318, 0.3970536902623138, 0.5267261402400154, 0.6253139538603372, 0.696541695221875, 0.7615326777534519, 0.8250161244760946, 0.8546525198028929, 0.7952713168583992, 0.6138815973927698, 0.33887744680252974, 0.055525916329423775, -0.1526205672662187, -0.2455207635574993, -0.2257612613468195, -0.14136417589496875, -0.07796346560781518, -0.1184046176944761, -0.2961174375323022, -0.5834688733246078, -0.9205990260160994, -1.2493793684817722, -1.5215623624805168, -1.7062321314215363, -1.7987075848960643, -1.810392811901411, -1.753152961113515, -1.638166642479112, -1.4801231948967195, -1.297224821923503, -1.1038877731463672, -0.9076438327859601, -0.7178353018656326, -0.545052696818883, -0.39679860778711856, -0.27603602577156333, -0.17899112701936187, -0.10174665980583089, -0.043863215826521924, -0.008092445801619426, -0.0006380208723445761, 0.02581751184577428, 0.06126652659504402, 0.08694560909602474, 0.08826064099184155, 0.07600816547751003, 0.09048325132799766, 0.1700068904262795, 0.3121355129350171, 0.4742695307470415, 0.606265798912187, 0.6766342395824964, 0.6890392482502093, 0.6795650858392027, 0.6873077118330191, 0.7075139461340706, 0.6884976788336737, 0.5762717697943507, 0.36504048036213294, 0.11299581813990978, -0.10538339091135318, -0.2412616303826065, -0.2791991534824294, -0.24638332040121758, -0.21600088348801955, -0.26827622940642554, -0.44002791844076483, -0.7104414398753478, -1.0229216912446848, -1.3190845353515472, -1.5561112893347249, -1.7106882730373996, -1.7827205492077498, -1.788498717188154, -1.741775068638831, -1.6481835212718285, -1.5155495285952019, -1.3579983941274922, -1.188234194544257, -1.0106400026440696, -0.8308930505810882, -0.6589686891645812, -0.5028927972218908, -0.36859469959204405, -0.255875207437407, -0.1592156586428931, -0.07515039971324969, 0.02765206904034957, 0.062034499610605136, 0.1041340334609387, 0.1423455480603322, 0.15444520157658762, 0.1297132826438391, 0.09528309404478758, 0.10920321648424357, 0.21404130030933327, 0.3962663367238997, 0.5935197371918868, 0.736429811138026, 0.7846926551610529, 0.7430507425375489, 0.6579500711336882, 0.5919376113389382, 0.5687794909563496, 0.5533869321201623, 0.48695598508658444, 0.34044269716369596, 0.13863244551831433, -0.06626392604624119, -0.22662610353310414, -0.31565352159188964, -0.34217670760732777, -0.3621195558508521, -0.44404003690048727, -0.6243139514022958, -0.8849303567308031, -1.1754701570807338, -1.4385077552497367, -1.6340529789916463, -1.7484303485166162, -1.7892687870234694, -1.7751418154421512, -1.7242375474171192, -1.6414213055176823, -1.5279678559633352, -1.3929794124000492, -1.246778954202168, -1.0905008287848457, -0.9249211842600488, -0.7582076647501362, -0.5978574057311731, -0.4510595562225228, -0.32271007835089344, -0.20900286781603455, -0.10286204900248495, 0.061881630932531254, 0.12130852834411152, 0.17543299860294864, 0.21049458598756549, 0.2037394165203123, 0.15426278976830343, 0.10856364158113216, 0.1376412351015742, 0.28138371971397746, 0.5126410124545854, 0.7512316242898924, 0.912595244757205, 0.9489247024359907, 0.8663145240253289, 0.7165060337942878, 0.5716936289465818, 0.48013138752483897, 0.4304627144646591, 0.37096928930623824, 0.26043709491997746, 0.09746673026224147, -0.08419687619874879, -0.24353754776381004, -0.35433733683896046, -0.4187501714627014, -0.4771394929535099, -0.5824318309596695, -0.7708738770231857, -1.0294028268862032, -1.3125481174965314, -1.5643905176669313, -1.7373821439774053, -1.8178361033046986, -1.8256530710026249, -1.7855511394207981, -1.7190576695945754, -1.6347844911884175, -1.5300378203485006, -1.4092787036497223, -1.2814417249514574, -1.1453582413411358, -0.9962359511027771, -0.8383310500530055, -0.6777069656342851, -0.5212157306826447, -0.3780605398712999, -0.24883122768568136, -0.12481501758545883, 0.09070396383780507, 0.17088485533115097, 0.2323855946587808, 0.2590290360109866, 0.23224770311234164, 0.16607130631578898, 0.12441922569605202, 0.18269231469752406, 0.3701340317310327, 0.6458596323104391, 0.9216064637111359, 1.1062434559043697, 1.1430873253426577, 1.0350632728549902, 0.8361587896559556, 0.6238069388014627, 0.4622435660675291, 0.35834652879363854, 0.2702903846432683, 0.1555870783052216, 0.0027034722715678027, -0.161499218990277, -0.3033436548752216, -0.4072444128619555, -0.478908798306556, -0.5481495099206359, -0.6556767878058889, -0.8368104259408365, -1.0886905313048023, -1.3706087111810343, -1.633348179851385, -1.8212869062311021, -1.9014680042158567, -1.891790102828613, -1.8278240892742403, -1.7384135682951571, -1.641847932652568, -1.5363848894377663, -1.4202758773248338, -1.301766450357817, -1.1804012961096624, -1.0466629000756698, -0.8995041513179692, -0.7423425326739003, -0.5798934760501552, -0.4237948382407616, -0.2801118990414922, -0.14126027997697405, 0.11134930376069437, 0.20593115470077314, 0.2697847113403655, 0.285391565191191, 0.24264466976360505, 0.17197861536986542, 0.1491875406290255, 0.24401871811162498, 0.4676495119281709, 0.7710247438720528, 1.0713729215050123, 1.276858227815684, 1.3222156461066654, 1.2058520409094304, 0.9811187600449612, 0.7260870558403044, 0.508948276729706, 0.34486081164729626, 0.20484058782053405, 0.055533581042925016, -0.11161503008655431, -0.2680567288248469, -0.38975764381166156, -0.4736004678519405, -0.5309985869793263, -0.5858346340006695, -0.6750336885781206, -0.8317857458895555, -1.062217864645089, -1.3315300197744797, -1.6029129060099567, -1.8292588798359468, -1.9561735544903731, -1.9711984460575067, -1.9049783862908936, -1.7941096442859874, -1.6734244228365929, -1.555866478846574, -1.4364343339068177, -1.318382228434276, -1.2036433124007755, -1.08099681686003, -0.9439301479229256, -0.7927249654942623, -0.6285305021589863, -0.4623654075757671, -0.3053668896968462, -0.15369388021872687, 0.12195664643762472, 0.2234312637967, 0.28550310301634285, 0.29122122874834594, 0.2411546741140264, 0.17962879804633355, 0.1858866692647495, 0.31525530165291726, 0.5606281344372325, 0.8727214844833608, 1.1782109473606404, 1.3883389375923474, 1.439157453388043, 1.3282603250345077, 1.103758246124721, 0.8365776901947249, 0.5875554367978337, 0.37397195903007713, 0.18050633336899363, -0.011896413327020459, -0.20077767917879466, -0.35891984039377584, -0.4729301982157647, -0.543990042840812, -0.5823889347157727, -0.607330558594228, -0.6583297073315553, -0.773654603757554, -0.967436410489347, -1.2135474943818938, -1.4843604647471191, -1.7481363854352703, -1.9443236773968264, -2.024608807110656, -1.9935947069511326, -1.882983027411612, -1.7378829072158848, -1.5957529917558184, -1.4626547189398533, -1.3371299652442021, -1.2208163525192415, -1.1033923575071756, -0.9741725753214477, -0.8300494165880185, -0.6680728866022259, -0.4955710489375142, -0.3268874241345493, -0.16369148546326862, 0.12244561383796815, 0.22341250138461066, 0.28121283002001596, 0.2815638014812417, 0.23533230192439358, 0.19415289079503348, 0.23181474526005338, 0.38786245247148327, 0.6431800237852745, 0.9483758875837616, 1.2358236862710423, 1.4268057590932288, 1.468787180636276, 1.3622882488557995, 1.1529876917635384, 0.903595122648466, 0.6582519118779826, 0.42611694920007004, 0.20047391673736245, -0.021554630746024775, -0.22905774853061234, -0.40493960879946256, -0.5396938077042043, -0.6223293185071456, -0.6510255895071763, -0.6396490070165048, -0.627080600701252, -0.6698566340793756, -0.8022016125587742, -1.0162887270099734, -1.292131164701779, -1.6003294520434588, -1.8722384092014408, -2.032688656054962, -2.06085014430038, -1.977418340873741, -1.8247161812365686, -1.6569806264687135, -1.5007944771882988, -1.3589405976423266, -1.2333902885176826, -1.1154514969172402, -0.9915330065619307, -0.8550947662431212, -0.6988856962124675, -0.524436118124976, -0.3465689806431971, -0.17276866183275513, 0.1150711463107993, 0.20993965557758543, 0.26273206954088374, 0.26323505449970885, 0.2297932896595433, 0.21384408814047784, 0.27861339972445415, 0.4513235255504479, 0.7056890791192337, 0.9913052501047599, 1.2452841967086863, 1.3992729513632867, 1.4121867167782531, 1.2942597418195239, 1.1017331076778085, 0.8946906277520049, 0.6933137108314971, 0.48836529901820774, 0.27406810101255624, 0.05275323262016902, -0.1701511404685366, -0.38790394212229373, -0.5805035447658263, -0.7078474851211295, -0.7477336340360601, -0.7049750350822566, -0.6118311683408336, -0.546946788073082, -0.5834881052363369, -0.7447585105384089, -1.024232696152968, -1.388109821655481, -1.7472974344815708, -1.9961976147475988, -2.092575427103428, -2.0524432636337795, -1.9115111836043661, -1.7276807968962427, -1.5468040892626465, -1.382925997129001, -1.241928756348235, -1.11861603414604, -0.9977552295418016, -0.8691815009230935, -0.7215376278175752, -0.5495816780494147, -0.3657039940266968, -0.1821582009520169, 0.10340479732636412, 0.18943593460917388, 0.23748196183724815, 0.24141535402334535, 0.22409992163361125, 0.2320254924263986, 0.31509314221679935, 0.49071384290699477, 0.731098704940494, 0.988219344729181, 1.2022551094179608, 1.3106931224211984, 1.2832058090952574, 1.14730862211315, 0.9781282746658063, 0.8332376676368034, 0.7090907782296297, 0.5773323071516046, 0.42222257772762767, 0.23112583482741617, -0.00936487475606339, -0.2910494548301905, -0.5642960978409025, -0.7588165261525742, -0.8295094694914238, -0.7702747261550088, -0.6107830120089897, -0.4396143096013433, -0.36692910577547183, -0.45867598351903316, -0.7262361776283028, -1.1331857933988658, -1.5727614933716345, -1.9111920836091583, -2.0804138054509003, -2.092380745203695, -1.9797889756918192, -1.7944072028570013, -1.5941007669097738, -1.4079966198459521, -1.2492291988732984, -1.1174760346830326, -0.9977101351818886, -0.8766088014286827, -0.7389152811691723, -0.5725399842216614, -0.3854330683167203, -0.1927369951385338, 0.0904448316002536, 0.1669649587108771, 0.2107309297911664, 0.21847861258324944, 0.21479366984583406, 0.23937889007033647, 0.32906082415170435, 0.4940831925805904, 0.7086188769634061, 0.9289638072997827, 1.100056305051202, 1.166444882336099, 1.1101063563326334, 0.97731561260268, 0.8521203405733455, 0.7803607031694613, 0.7446763045630215, 0.7072359632000881, 0.6366541345800878, 0.4918379598636915, 0.24126681665456473, -0.09767522158170899, -0.44788010812200646, -0.716817741725735, -0.834387158915065, -0.7728579560017718, -0.5822049319105619, -0.3566138885529919, -0.2110570921951196, -0.24001710934287662, -0.47875971863889816, -0.8967645232122236, -1.3823715110969599, -1.7893448673518906, -2.025610636801733, -2.0912987947307577, -2.0176803670948837, -1.846277275044792, -1.6371919303018188, -1.4339062619812393, -1.2591400215909732, -1.118527425898087, -0.9987065415345932, -0.8839839136754197, -0.7556659628209959, -0.5956387213645987, -0.40654986175185626, -0.20467625615656418, 0.0771603797996852, 0.14421889581397118, 0.18393040761591262, 0.19347113474925132, 0.196642723159097, 0.22703404565867225, 0.3116487736531793, 0.4554438953359498, 0.6349379352119561, 0.8140252356927838, 0.9472978163377704, 0.9883015464305033, 0.9319916630681526, 0.8342818373545394, 0.7683519995035463, 0.766179424560556, 0.8105049203573272, 0.8612088560200294, 0.8708032993430062, 0.7789763757111247, 0.5433720340601448, 0.17986735621745617, -0.22304388207025364, -0.553016343026878, -0.7217485259199571, -0.6872403771204516, -0.5078223138742345, -0.28873571743398785, -0.131520056685441, -0.13001172282025145, -0.3347674572756933, -0.7287985735152963, -1.2149113035856536, -1.6542382931415398, -1.9403041492528192, -2.054129872054855, -2.0230129517241116, -1.8773829340597263, -1.671114601736596, -1.4588463190694254, -1.2736394956148724, -1.126544904993686, -1.006455230915621, -0.8962866172083362, -0.7747807622682331, -0.6194382269275892, -0.4281087681427085, -0.21705806052301926, 0.06313563418106936, 0.11979471673078212, 0.15422867949594463, 0.16226279121550935, 0.16468394383558796, 0.19000117874099964, 0.2587749701790219, 0.3734577635928887, 0.5147959789235278, 0.6556943394228527, 0.7620623046357659, 0.7986648820005203, 0.7695246613143788, 0.7265644333128984, 0.723086395311844, 0.7805864586160471, 0.8854553835896555, 0.9968992261827874, 1.0624064654935397, 1.0176709344616146, 0.8115438088805148, 0.45991301791269507, 0.05500607726411767, -0.27422268997014465, -0.4454200078802153, -0.4509906429200778, -0.3423790335125883, -0.19346733340680994, -0.08003254349369919, -0.08376223035171688, -0.26668949536460196, -0.6252810842099686, -1.082879227385759, -1.5221174166182898, -1.8368700593801128, -1.9883112797662978, -1.9963968966069638, -1.8832530913536174, -1.6911301648593755, -1.4792577340272435, -1.2907608016771026, -1.1417565818894522, -1.022519190670267, -0.9149996941346411, -0.7968258207561327, -0.6431581559120277, -0.4484534093978045, -0.22862491214159658, 0.04725022381865988, 0.09081923915137521, 0.11678700926753678, 0.11953161923974542, 0.11556089554312159, 0.12830758664367387, 0.17412340206417284, 0.255631634254216, 0.35911018936006106, 0.4667888354832606, 0.5573920757297844, 0.6068896705525122, 0.6216536851291155, 0.6407267022911609, 0.6937513144391481, 0.7924012656117126, 0.9261920759847515, 1.055450803710014, 1.1357236009617588, 1.1121528677392745, 0.9389982445826685, 0.6309085817790692, 0.28153830605310404, 0.005854117698188248, -0.1377237640841543, -0.16234488689709053, -0.11248348343552887, -0.03978323633889856, 0.007611015342791452, -0.0261023574777292, -0.20402977101635675, -0.5381100242823071, -0.9671267447219233, -1.3937363792810413, -1.7209163531992242, -1.8979862223970185, -1.9382015339090852, -1.8588240927720954, -1.6898836649990494, -1.4887553225936654, -1.3052987223261405, -1.1600231585650957, -1.044203162326917, -0.938691772962998, -0.8208634157231437, -0.6657105778206026, -0.46639525921017216, -0.2385435112448802, 0.02848665237306353, 0.05480741409755699, 0.06766925426488862, 0.06140894560237683, 0.04855143025496408, 0.0468526145728898, 0.06828945516879029, 0.11597124073004061, 0.18256893214328676, 0.26127313667244606, 0.343662297494012, 0.41426275331780354, 0.4765862222145876, 0.5522640272166537, 0.6464140586061586, 0.7578212194247086, 0.879285387858829, 0.9851461116947207, 1.043990649597272, 1.0162115960684814, 0.871586278837573, 0.6246714274443307, 0.363251672379064, 0.17644367478378056, 0.089194518185467, 0.08061311515779325, 0.11212327422782241, 0.14718755030746405, 0.15440393418007248, 0.09305495774777525, -0.09872545805213116, -0.4365086301409837, -0.8526380308565471, -1.2641903214170582, -1.5927937215506651, -1.7845950325940807, -1.8482486370425033, -1.8001000941341365, -1.6600158698203444, -1.478759168430078, -1.308880538211663, -1.1739580630061812, -1.0652866135641175, -0.9624954808407366, -0.8433233369344613, -0.6844721135063421, -0.48001652440960174, -0.24572126215589568, 0.005187701621061604, 0.009252498123288756, 0.004533196534217025, -0.013296285436871625, -0.03476975971144726, -0.04795742002885432, -0.046618138804540916, -0.028154936331584675, 0.005361194591510642, 0.05642524689767216, 0.12936990269327742, 0.21694538218749654, 0.3168705739890662, 0.4329390720549415, 0.5483026465667512, 0.6496996016376589, 0.7369527455444307, 0.790204717643887, 0.7855168508207417, 0.7241392418804473, 0.6069414074981895, 0.4452299300986511, 0.2965989473498929, 0.22071732244770326, 0.21797734704935515, 0.2535818247251899, 0.29626809567536405, 0.32791685402038384, 0.32888178947139163, 0.25718282417223165, 0.04914414417377276, -0.311963754702862, -0.7345394210507417, -1.1330145624524777, -1.4528645527379611, -1.6496887937787992, -1.7288888998070246, -1.7064247668142027, -1.5963411761970896, -1.4418085748599374, -1.2934689886029132, -1.175082921167418, -1.077138834517065, -0.9782919098715841, -0.8573526758781642, -0.6943828472239666, -0.48605485276008326, -0.24861238597896373, -0.024174294455669616, -0.04773497440234186, -0.07348555953804775, -0.103604766589945, -0.1316819254761268, -0.1507932324913725, -0.16027650866945978, -0.16078705238874522, -0.1533273647422707, -0.12928163680936272, -0.07271904369927544, 0.01709745903666078, 0.13337092871417405, 0.2674962760196219, 0.3880015836311255, 0.4690793359545437, 0.5114924145237358, 0.5027372585720818, 0.4269918285926347, 0.31421246376367395, 0.21104948176068417, 0.1325478380591925, 0.1013923431257935, 0.14432405025075606, 0.23533220543891165, 0.32712494373977896, 0.3963519547883603, 0.44803465744827414, 0.47325864542262597, 0.41624930442232977, 0.19882284465297728, -0.18669670778136155, -0.6156322963044453, -0.9968854798799909, -1.3009596700273054, -1.4960907136463408, -1.583821259088843, -1.5798209070827778, -1.49732235583601, -1.3732799981184212, -1.252532278271463, -1.1553247828248518, -1.0704953542497948, -0.9767612828006187, -0.855305635178686, -0.6905338378609684, -0.4820959007145761, -0.24633606095906468, -0.05919433616904175, -0.11455994531160853, -0.16313525839721785, -0.20499691614096593, -0.23700531216404744, -0.25621982782406993, -0.2665526367709933, -0.2728564310192108, -0.2789514673916566, -0.2776609576902814, -0.24691350028864456, -0.1750541959224624, -0.0662668099233081, 0.06354191610833902, 0.1742916267559958, 0.23346751841988206, 0.24089733212422937, 0.18809123627932844, 0.06502113115130338, -0.08271129286638561, -0.18540324150306028, -0.21618765729028247, -0.1638208812632969, -0.022106266282819594, 0.15726342096751017, 0.30704114728461346, 0.41108666106222463, 0.492721139584093, 0.5486298468139078, 0.5112270323700108, 0.29748279058633614, -0.08451746151660722, -0.49747482915033253, -0.853748592971923, -1.1393024660104818, -1.3288776246287124, -1.4184254395791647, -1.4239630940596593, -1.3635692436309288, -1.2708266248963216, -1.1811876672547548, -1.1077227632483575, -1.0375541010004456, -0.9510571597406294, -0.8327048743477607, -0.6711252624909896, -0.46827574033131336, -0.2394678426437066, -0.09634916617388124, -0.18463442528198346, -0.2558359955665878, -0.3083239373395258, -0.3425069891187556, -0.3581326443618209, -0.36144934024273184, -0.3615460551464523, -0.3677891030314698, -0.37964345391226617, -0.37606903939590924, -0.3379523337002049, -0.26121072437752935, -0.15637547553299094, -0.06579776647363471, -0.029885157245068777, -0.047823975334373436, -0.11895831031989507, -0.25508757499468365, -0.41410386159317253, -0.517419344154204, -0.5337922144511428, -0.44152574011986784, -0.2379529034424976, 0.010307413243333199, 0.21864884334802237, 0.36461118391179714, 0.4738164987027322, 0.5491785135689147, 0.5295501747239879, 0.3369055132914256, -0.009197802622012875, -0.381627999481323, -0.7068999677990584, -0.9741632496847107, -1.1544501346789164, -1.2377425490050602, -1.2433301132542565, -1.1985663454405586, -1.1351594389422635, -1.0770808080335084, -1.0286581048905408, -0.9753655694861914, -0.8999335122963403, -0.7901409812854832, -0.6379753032934838, -0.446494432718961, -0.22916786072865078, -0.1308541140627241, -0.24926781979430254, -0.34101306958380023, -0.4029316375532499, -0.43837243521630054, -0.4488476602386159, -0.4415792473501498, -0.42825308158999975, -0.42255873919723436, -0.4341604746920314, -0.45146052425795374, -0.45175382772896866, -0.4210387219248772, -0.35771975377021487, -0.2934718861077369, -0.2777886714656149, -0.32182155540285945, -0.4119434642764208, -0.5465959114404593, -0.6962609879899918, -0.787299611033412, -0.7903294099719645, -0.683574428041477, -0.45437724433975013, -0.16627996979719717, 0.08986601333836332, 0.27751041817875255, 0.412668670286208, 0.5011854079646415, 0.4974267200414659, 0.337790709867074, 0.04766154580118715, -0.2722013844792221, -0.5653585739708725, -0.8118012261440081, -0.974736546339442, -1.0427553723256282, -1.041178690094087, -1.0074431973490519, -0.9707725488676173, -0.9428949008428584, -0.9198760797065442, -0.8863057993418125, -0.8268372549933337, -0.7312899113239223, -0.5940062197114179, -0.4184636829671622, -0.2161191512817835, -0.1590375362148253, -0.3019094288056093, -0.41086582190495813, -0.48104323188805653, -0.5172971886439777, -0.5220293781627929, -0.5025934618111806, -0.47290803075506427, -0.44898703015514113, -0.4471533409228029, -0.46943431431100013, -0.499815905587329, -0.5168359978094917, -0.502630381308242, -0.4699875446190872, -0.46898129037186786, -0.5313227555340101, -0.6468151396330588, -0.7934160946107923, -0.9330758978882634, -1.0030069724478712, -0.9811891103390625, -0.8624013080064832, -0.6286555394205484, -0.3254950644818805, -0.037641196799146036, 0.18377814364292352, 0.33988627294152046, 0.43844651969326826, 0.4462514086272235, 0.323422867783371, 0.09276873493830547, -0.17529721289714384, -0.4337602553172001, -0.6490372054328181, -0.7836319440851989, -0.8303704028705365, -0.8209201152218065, -0.7986323339788434, -0.7862611341641913, -0.7849584097239106, -0.786270435187782, -0.774740734030772, -0.7353378633577212, -0.6584562696887533, -0.5401222483147091, -0.38410774603212827, -0.19996252473812776, -0.1791614420411132, -0.339656998780478, -0.4621956179760632, -0.5396631830167313, -0.5759751666175701, -0.5740626930822262, -0.5416164277698079, -0.4942899900884139, -0.44953510530069746, -0.42657513706751826, -0.4373062198037745, -0.4767919951615472, -0.5260416510215176, -0.557594522393088, -0.5638278490322752, -0.5819435681749712, -0.6534574900707077, -0.7826292095889104, -0.9442698703111466, -1.0892958670831983, -1.1516288434747655, -1.111785443034118, -0.9817546127316263, -0.7493424337815046, -0.4415002429269256, -0.13321461385319538, 0.11583809224277178, 0.28942073030016213, 0.39219932659563594, 0.40570167601975277, 0.31541771547071884, 0.1360954124941214, -0.08403732901364719, -0.30121248312539134, -0.476071863495531, -0.577391568476602, -0.6033700118875563, -0.5912139976411729, -0.582977065505654, -0.5913836187969751, -0.6115397456700415, -0.6346861522839741, -0.6455570554176892, -0.6281492250121382, -0.5725009089786577, -0.4758298041911813, -0.34231293339914587, -0.179826031096638, -0.1913885882150099, -0.36275573368530006, -0.49522969553648455, -0.5787727407073368, -0.6138883789915179, -0.6039449011732648, -0.5573998572380577, -0.49132088991712497, -0.42468052290308034, -0.37682706365472635, -0.3644164834659693, -0.3927168588139096, -0.45178492983820134, -0.5147121952732102, -0.5610666082689584, -0.6087185253346701, -0.6919990326045834, -0.8243492301063925, -0.9903055590536178, -1.1449114859456389, -1.2189907570719476, -1.1821052049914986, -1.0506191383341055, -0.8229955028601089, -0.515862183484204, -0.1930351167807005, 0.08149463222847941, 0.2729921377302224, 0.37845016060238096, 0.39721578476013486, 0.3304560914263305, 0.19146314942994594, 0.014565373154063628, -0.15928592347354417, -0.29212434740147236, -0.3605839971619907, -0.37102283239205547, -0.3642766943262201, -0.3722271165663359, -0.39694895730546387, -0.4331397887186654, -0.47465793447653154, -0.5061935237358677, -0.5101380486599693, -0.47601857949748666, -0.4021676135871992, -0.29311945541006007, -0.15546206449981717, -0.1963121800603594, -0.3726234038516739, -0.5113896572149025, -0.5991223709365321, -0.6309054029216097, -0.6104615840727663, -0.5488014608456858, -0.4637164816946536, -0.37486929247725687, -0.30167235103237666, -0.26132819685354886, -0.26410642058900163, -0.311934412469125, -0.3889004229592278, -0.46791880125558005, -0.5480547108221883, -0.650294852497981, -0.7875832183606876, -0.9542184820170334, -1.1155356371804968, -1.208577061946629, -1.1887816769902773, -1.064746009139752, -0.8449683459571312, -0.5439255092022153, -0.21517720721470393, 0.07463609790931805, 0.2811076001649959, 0.3925911825177085, 0.4178037443684104, 0.3683666406930807, 0.2609427717228454, 0.12254644121751565, -0.007318625869567721, -0.09956330209789675, -0.14076106244769213, -0.14666538571484333, -0.1533707289421981, -0.1765773051899184, -0.21314812801890895, -0.2621447855193414, -0.31904985924102336, -0.367361932984568, -0.38914840855516414, -0.3747542185723716, -0.323410520358649, -0.23934621855662042, -0.12821518921483316, -0.1945133715389886, -0.3703819561877546, -0.5108659396664506, -0.5995172124438165, -0.6261161580660813, -0.5931406634592811, -0.5143521371324445, -0.4100061872634485, -0.30034463002611156, -0.20388993942705802, -0.13757410880300316, -0.1122966913252568, -0.1346314273158346, -0.20398691899909685, -0.2994277030164983, -0.4031781488871816, -0.5218404999425446, -0.6639186059273995, -0.8286113947132874, -0.9916751314743549, -1.0994509479647743, -1.100893713888255, -0.9916004865666833, -0.7867807144878991, -0.5044348207425026, -0.19006087443157768, 0.09416545825395481, 0.3032331762020172, 0.42053767562339484, 0.4558594318334459, 0.4253056174402507, 0.34614548147830715, 0.2460722563385349, 0.15694661395492693, 0.09622319184445077, 0.06852216946270748, 0.05467022020815147, 0.03254782954989338, -0.0024673843941558113, -0.049949241758827326, -0.1115850163489491, -0.18035495891792833, -0.23919337158146514, -0.2732235767465085, -0.27540380437169054, -0.2451012119674992, -0.18523815284450662, -0.10045724839569158, -0.18772554960718854, -0.3574977900597438, -0.49275960090016746, -0.5763992743199277, -0.5956015171154554, -0.5505335336526488, -0.45484723884000827, -0.3310091243712074, -0.20213740117239806, -0.08629304410633046, 0.0016819972054767766, 0.05062949101244551, 0.054156575386933865, 0.007095996521453962, -0.0833495175925297, -0.19486106918713056, -0.31775636112073613, -0.4542725288230422, -0.6045798731812287, -0.7555875928988297, -0.866107837090595, -0.8879915057206587, -0.8054934111916727, -0.6325066819608406, -0.38936424474222736, -0.11354788987899467, 0.1444319739030899, 0.3436883382656386, 0.4667462670688271, 0.5174799462783557, 0.5080853521467101, 0.4563854381580207, 0.3896069844695625, 0.3283863556808484, 0.2827969670828964, 0.2523841990950951, 0.22478140424123338, 0.18983996808200107, 0.14318841166564833, 0.08231794975327962, 0.008761825437077878, -0.0663577749619666, -0.12830281821065243, -0.16837868915242887, -0.1831737006914348, -0.17149865787532687, -0.13410171514071156, -0.07414240234491863, -0.17674371786501406, -0.33436484935876365, -0.4560224515653616, -0.526832043992768, -0.5355317210097299, -0.4801432986918088, -0.37099632555121975, -0.2301012086710807, -0.08397589001867407, 0.0473384283364685, 0.15166309527033295, 0.22005959823161025, 0.2455551739734238, 0.22270780599582957, 0.15120673987222077, 0.046828577976454414, -0.07060041348128025, -0.19273445604842782, -0.3176014602285261, -0.44097315975333534, -0.5390364074504645, -0.5734954085126648, -0.523572471124537, -0.3947415777970127, -0.20386433036749216, 0.021066224641410055, 0.24212406245303014, 0.4246994936679429, 0.5494524558237964, 0.6120712848228888, 0.6198493171279557, 0.590579609964687, 0.5452508546905888, 0.49560517129994675, 0.4483574984071154, 0.4057934619049482, 0.3637136054881354, 0.31410613313925073, 0.2515000987305498, 0.17620943775732323, 0.09519118237278167, 0.02107437717145434, -0.03726339636058258, -0.07838227614859518, -0.10182074038169707, -0.10552023042256466, -0.08788732753822118, -0.05032508054401927, -0.15960314569436823, -0.2986436893386519, -0.3999362504566184, -0.45127466147999484, -0.44551431667650243, -0.3813811672888744, -0.26493453405703193, -0.11303205143812263, 0.047098995540529105, 0.19123058786841013, 0.3073948889420061, 0.3904355860007047, 0.43508690927960686, 0.4351951824581723, 0.38766745501283023, 0.30120775799697197, 0.19578738022729542, 0.08923456447827698, -0.009817392424300134, -0.10038083770023924, -0.17459625429478062, -0.2082712441477596, -0.18188950760075254, -0.09317498258385594, 0.04747414005331997, 0.2209833189084123, 0.40029612121672764, 0.5570838148665238, 0.6718637803993961, 0.7360647974505766, 0.7538986845432466, 0.7382136903994947, 0.7003136074977074, 0.6475481324255629, 0.5889171126862708, 0.5303440962300695, 0.46872155535625554, 0.3978237676056098, 0.31700272019298414, 0.23166512220830812, 0.15143441621724088, 0.0851540935231133, 0.033804129843938414, -0.0061804888655291575, -0.03524931780457811, -0.050550082937106826, -0.04889528830238959, -0.030116586184948897, -0.1339111280659618, -0.24811957166393364, -0.32558985162533105, -0.35476356680344434, -0.33139838059510796, -0.25767816444747377, -0.1391177325805825, 0.014021849251428528, 0.1804935472825606, 0.33430820106056475, 0.4600723560912467, 0.5536473940509001, 0.6128470901032134, 0.6328776224249417, 0.6091465242158, 0.545591207803891, 0.45817199182055773, 0.36744533348399133, 0.2892795324825232, 0.2275970710321696, 0.1810463112260952, 0.15954532818874087, 0.176846595256202, 0.23826934692482524, 0.3390309010943746, 0.46702865410504596, 0.6039851758659143, 0.728696896559049, 0.8248304591653794, 0.8824361948989551, 0.9015200777758305, 0.8857777276739761, 0.8406917921727933, 0.7751757528308421, 0.7000728288402793, 0.6195568983626921, 0.5320300038509831, 0.43821310796503776, 0.3439550598698426, 0.2574443162917144, 0.18559156618659095, 0.12960163285404502, 0.0844719209532788, 0.04542616316065822, 0.012540550014051867, -0.010763189440408176, -0.02046790772334356, -0.01530139779539877, -0.1005808549508258, -0.18491330925823002, -0.23722397759496228, -0.24507749010444346, -0.20432420324700762, -0.1199756398753009, -0.00027705360648431056, 0.1466046619315825, 0.3077993556758355, 0.4631611585079445, 0.5953992025535685, 0.6974620306444772, 0.7671545311408945, 0.8015573506119996, 0.7970753719629646, 0.7547726538208153, 0.6866839790690207, 0.6120011398250037, 0.5497737329880589, 0.5096535756491382, 0.4885572958739258, 0.48520262236182915, 0.5055503619743805, 0.5534856025626681, 0.6281681129114224, 0.7223977987234728, 0.8241035778690777, 0.9188575576316372, 0.9928732440440177, 1.036017936402031, 1.0429064416247287, 1.0121444069035344, 0.9499802376236622, 0.8666811054162067, 0.7707046919158829, 0.665050709143761, 0.5533346100223454, 0.44335180439276756, 0.34500091189329823, 0.2648390637503988, 0.2033649889431852, 0.15555112246395766, 0.11422677407317712, 0.07534018265241552, 0.039791173169141346, 0.01166396417256468, -0.004406304621426101, -0.006822033688186716, -0.06380435899086306, -0.11545502504033156, -0.14200955209557098, -0.1309934079730372, -0.07631643144061782, 0.016932930460685985, 0.13759314690891158, 0.2758404661750859, 0.4232619910164914, 0.5686202811866689, 0.6991927604106497, 0.8057469981888774, 0.8831625997237201, 0.9279399063891345, 0.9377289736513393, 0.9129543142570604, 0.8621702858556199, 0.8022926033766257, 0.7522245158941946, 0.7258538185585472, 0.7239667084031931, 0.7395509194372263, 0.7707288061036578, 0.8172744268771559, 0.8790983958912936, 0.9517623470712242, 1.0270721605989024, 1.0947101538679391, 1.1430872533481324, 1.162969714295583, 1.1469205744986024, 1.09494902657221, 1.0137313762926683, 0.9116903513361134, 0.7950959935170724, 0.6689996893382281, 0.5423229604502999, 0.4269304108025723, 0.3321706140363014, 0.26014541517510803, 0.2067726507835, 0.1642387819294532, 0.12497584079315827, 0.08598967435770072, 0.0491801341266471, 0.01931001743329589, 0.0012699544266397181, -0.003675287194829899, -0.028014941920016006, -0.04766110331988284, -0.049377767289123346, -0.02231221311349742, 0.04107740789545217, 0.13817444802391457, 0.2574460583092191, 0.386829317915015, 0.5184181538266476, 0.6465051579474135, 0.7651550854023674, 0.8674669569758593, 0.9466243256300001, 0.9973799206670576, 1.0169401463187238, 1.0058439640503238, 0.970217641415328, 0.9239332263842812, 0.8843658856964722, 0.866439956878365, 0.8757662808938244, 0.9058390892269481, 0.9491376264006032, 1.0009455482009622, 1.0583082100067844, 1.1176116218300818, 1.1726448207487237, 1.2154622417917584, 1.2375546375532709, 1.2314769898055775, 1.1922764456688635, 1.121606919984251, 1.0241446537584042, 0.906638416622811, 0.7755720197121955, 0.6394651270337365, 0.5100055540698419, 0.3980729840355885, 0.30952166204877757, 0.2444964990227117, 0.1976407649080307, 0.1593260867597421, 0.12201335855917235, 0.08400748919632176, 0.0478719639482481, 0.018614628221435114, 0.0011642802847262794, -0.0035242337188272537, 0.0031597795124251584, 0.011008162617546214, 0.03055724496458833, 0.07059913400682716, 0.13784864968781543, 0.23186651622070165, 0.344187014737178, 0.46307534847148146, 0.579849435709111, 0.6903414934631987, 0.792166554473253, 0.8820956798643886, 0.9548583858183437, 1.0046109494959163, 1.0275516410263785, 1.02375146987609, 0.998653504559233, 0.9634997597750331, 0.9334173450726673, 0.9221333137227093, 0.9371098665559678, 0.9751575557143943, 1.0269763815699586, 1.0845427778699972, 1.1413816636524794, 1.1928870337113444, 1.2334609297756858, 1.2566999480738705, 1.2566993531793291, 1.228575414979208, 1.171090507733476, 1.0861241512873565, 0.977791152134623, 0.8525352349178345, 0.7179980317149018, 0.584442057486685, 0.461897728560019, 0.3574223518217122, 0.27577338819173014, 0.2182640312226401, 0.17856114216029473, 0.14555385827696846, 0.11221075173123664, 0.07746937900824657, 0.04376937918068699, 0.0161758553099998, -0.00016255621985892757, -0.004090489589853737, 0.026829316125256426, 0.054639213307454124, 0.08894956241722607, 0.13747320837343271, 0.20489698935789027, 0.2905902722824736, 0.38954333822957443, 0.49359723583578907, 0.5948381083651607, 0.6889518012885645, 0.774364701099091, 0.8494062461456541, 0.9111076589023044, 0.9544911684431043, 0.9750014441237894, 0.9720367923335966, 0.9509915375199259, 0.9223024564527346, 0.8991524291832206, 0.8936700042487019, 0.9123448999556514, 0.95390834097376, 1.009776839601775, 1.070248435211096, 1.1267489196944231, 1.173056081541814, 1.2034345094081746, 1.212169765497118, 1.1950801616707682, 1.1505061039577975, 1.0803570463239207, 0.9879087163198857, 0.8781408919022242, 0.75675063689762, 0.6309933541151377, 0.5092538507478587, 0.39826142716884116, 0.30372615060213415, 0.2315375453256844, 0.1833093026913351, 0.15188940166708811, 0.12648489806654717, 0.10023955981719018, 0.07122912838996066, 0.04137278281296644, 0.01590362713302265, 0.00034467631107543964, -0.003664813602973828, 0.040742885993823016, 0.07962328127201031, 0.12066582734238528, 0.1710018262093876, 0.23408109929247786, 0.3084186737972767, 0.390557605797574, 0.47586031974928467, 0.558930616836697, 0.6358074214923447, 0.7049559139044712, 0.765232336706661, 0.8148415629365715, 0.8499948432087202, 0.8657617408895784, 0.8609795827669742, 0.840539330721325, 0.8145093480202735, 0.7950366282439771, 0.7931313239445648, 0.8140133633425991, 0.8561613974080611, 0.9117785156923272, 0.9706040486771895, 1.0234936730890631, 1.0635440055566665, 1.0849277871279606, 1.082576442376346, 1.0540220529542548, 1.0007066012136883, 0.9268470531766626, 0.8376251802432015, 0.7378470698802772, 0.6312848830755486, 0.5227155001546131, 0.4176656009330615, 0.3217754910446095, 0.24120954314794385, 0.1812845022217478, 0.14280414955837883, 0.11997228783001848, 0.10363722839588921, 0.08629312049150265, 0.06462304245313889, 0.04005177578098609, 0.017669112678716098, 0.0030645339089468634, -0.0017804847560341, 0.043369079374086385, 0.08410760721315727, 0.12428448618359063, 0.16932803949002492, 0.22220653467084373, 0.28181711061327663, 0.34569678753335886, 0.4110060485511572, 0.474398433632556, 0.5329129862083093, 0.5849365871073567, 0.629926361633608, 0.6668973401909789, 0.692997311714752, 0.7038121521178045, 0.6977385892447012, 0.6786244491315699, 0.6549257755698353, 0.6375709911157227, 0.6365682799321909, 0.6565899145209176, 0.6953766698911282, 0.7455350589775128, 0.7971396983278946, 0.8417810632266232, 0.8732679292423149, 0.8865668611818666, 0.8777251090276241, 0.8458597268180701, 0.7940817097496444, 0.7276061741475901, 0.6519938611718064, 0.5707925330100735, 0.4859773470451961, 0.39975713408252334, 0.31600324510088756, 0.24006334718321964, 0.1770145870670936, 0.13034132365021348, 0.10115740344105761, 0.08624000526711344, 0.07797267742724236, 0.06859530461496537, 0.05434402616978545, 0.03624356771545867, 0.018506915906971528, 0.005927368724378735, 0.0004736729875959821, 0.03527516162632222, 0.06889617387216296, 0.10133317606976898, 0.13530529781491588, 0.17299393436574248, 0.21424594107169062, 0.25795426523433357, 0.30259760819560916, 0.34588576885973094, 0.3856743788078016, 0.42048939411223846, 0.44996448502455527, 0.47397591958658813, 0.49067776190633505, 0.4969338940033937, 0.4912448354297121, 0.4760874429558591, 0.4573118571482667, 0.4432220740816185, 0.44195687472588036, 0.45751332296839037, 0.4877593243888226, 0.5262479747448463, 0.5648600396134025, 0.5971209835726955, 0.6186406305318962, 0.625902843463064, 0.6162748010987, 0.5897682983899983, 0.5493024847826564, 0.4994177911972959, 0.4445256019065788, 0.38715640842037974, 0.3282856117552073, 0.2687257857803887, 0.21108241401236905, 0.15897596328872934, 0.11528119769127089, 0.08253988843357733, 0.06278283938870387, 0.05440625395471765, 0.05132157517180626, 0.047193025440550346, 0.03912958738321443, 0.027726750028452478, 0.015765868557967692, 0.0065752550136423185, 0.0017114455626505525, 0.019487394428487678, 0.0384544799107768, 0.05678252763158428, 0.07520749896682483, 0.09476316250123054, 0.11563298538208702, 0.1376390234640961, 0.16038682940874002, 0.18258097579355662, 0.20292032660027134, 0.22053649566936737, 0.23504827948416898, 0.2466860119798231, 0.2546339534538899, 0.25728199276587255, 0.2538427138955741, 0.24540318114019968, 0.2349158222433197, 0.2268147012801911, 0.2257941328039998, 0.23422222435860304, 0.25089932630972617, 0.27190042906719714, 0.2926469982137596, 0.30954870660553074, 0.3204529268368251, 0.32361039428392036, 0.3176867274371309, 0.30283499734085023, 0.2806969915712188, 0.25387036740471836, 0.224783960616863, 0.19496399276568863, 0.16484662134148073, 0.13472500129489318, 0.10573573650313867, 0.07929376285253886, 0.05664183271677165, 0.03951673644669574, 0.02958434381567372, 0.0260243663246901, 0.025285885044968807, 0.023879874069589063, 0.020364939696053283, 0.015008146964804231, 0.009090670544440073, 0.00425605204167159, 0.001379307673420638, 5.510891837154826, 0.005123277443138685, 0.013143197381536624, 0.025591418920287448, 0.0421633092540315, 0.06063286689247589, 0.07837332102394105, 0.09404130141375376, 0.10782533981364605, 0.1207383126558213, 0.13357082017847452, 0.14518741743458713, 0.15396433023105358, 0.1581460808372101, 0.15639359412526904, 0.14889588636712128, 0.1366172766148176, 0.12030972367199244, 0.10096282755632041, 0.07971770144641328, 0.05693371075358788, 0.032352254459325804, 0.004942218684542141, -0.025750161954848264, -0.058557721224185955, -0.09181595198000092, -0.12518826571102318, -0.1593036864577879, -0.19317583007642014, -0.2237484612817865, -0.24900355726310577, -0.26945728992717427, -0.2869783571782132, -0.3019514736027138, -0.3130072651400551, -0.31984599722392754, -0.3231430591929813, -0.3221123494654594, -0.3138569784651194, -0.2961912618487893, -0.2699811869276096, -0.23905023148682558, -0.20768133225876328, -0.17774501224846465, -0.14826841304085772, -0.11793519322403423, -0.08695505394452947, -0.056558397333751266, -0.027634405541235373, 0.007041833324956083, 0.020350669313317964, 0.04271971826081542, 0.07303488955090762, 0.10727062984268021, 0.14073242468113736, 0.17067446035502715, 0.19715659631998286, 0.222113454313086, 0.24613468222879314, 0.26714660307019295, 0.28260628055587533, 0.2897560903129787, 0.28600917290963607, 0.2704437932967104, 0.24344292503731443, 0.2068659902580087, 0.16482762220640004, 0.1221029010780982, 0.08120115507261903, 0.04139267422742126, -0.0011190327528098557, -0.05045605183229241, -0.10751539858135774, -0.17062946959072284, -0.23811143337996563, -0.30892144098540314, -0.38010506604257394, -0.4456288039426805, -0.5001050537190213, -0.542775753143367, -0.577055254277088, -0.6054218274849714, -0.6267408282289029, -0.6395953173787701, -0.6441447331968301, -0.6394559524288023, -0.6219403276964259, -0.5882741295971391, -0.5393898705342277, -0.4809844399930067, -0.4199701834835653, -0.35989106906952434, -0.2999304805371118, -0.2384328803989873, -0.1757993877115396, -0.11424516918141693, -0.05579161698188116, 0.00462785953632739, 0.018594826928910187, 0.04549497469965896, 0.08352889633262767, 0.12790114725333138, 0.17276487324531487, 0.21395960414352763, 0.25087922178278377, 0.2854983063196785, 0.31799453620322404, 0.3462188836926916, 0.3674960051818706, 0.37835722073880335, 0.3741014499036298, 0.35038615755278585, 0.3053480492645164, 0.2425623532856751, 0.17204735552238748, 0.10539742503532162, 0.049866640652925474, 0.005246549893379406, -0.03625524306790998, -0.0862080103583004, -0.15286410511981618, -0.23826053188513657, -0.3391392139228528, -0.44942721793099427, -0.5614453388810285, -0.6656380693508261, -0.7527927501384571, -0.8192975552892933, -0.8691556079860077, -0.9082929953906415, -0.9376895097681367, -0.9545991449566179, -0.9574587264720205, -0.9456561514059857, -0.9173241373967674, -0.8697493907276934, -0.8030910028329904, -0.7224318947777467, -0.6349929753101458, -0.5455538834689729, -0.4546805733429123, -0.3613552289670682, -0.2665255106531525, -0.17340592152564718, -0.08479320535411382, -0.0022034712220318294, 0.006354397820233398, 0.030001400773587455, 0.06769185171875593, 0.11508335922893326, 0.16562168604607355, 0.2136333971284447, 0.2574988411085, 0.29864849702135843, 0.33777381987009486, 0.37350086396568494, 0.4030145452801898, 0.4210854715037343, 0.4192085095771212, 0.38787447008217146, 0.32240721625457497, 0.2285948399952198, 0.12344352535799734, 0.02729663473821982, -0.045095668683578796, -0.09074313359347397, -0.12059451740095681, -0.1554302680811847, -0.21526489586415057, -0.3105833506176558, -0.4389409130981646, -0.5875311554172699, -0.7402966921295918, -0.8822034755813333, -1.0008224556922005, -1.0900133343462666, -1.1534580785214184, -1.2002702516714763, -1.2347213097475758, -1.2536285327862382, -1.2530683704373027, -1.2325925770713795, -1.1928893646472445, -1.133044957090356, -1.0522842828017613, -0.9538647340835693, -0.8441354531658237, -0.7283131420511912, -0.608160217327476, -0.48381886284781417, -0.3572691028269401, -0.232882573372475, -0.11405572882772506, -0.013852922315443291, -0.017768217097388427, -0.005886480537640927, 0.0234525563973217, 0.06699937324865746, 0.1172468419218738, 0.1671027031596277, 0.21415091198702949, 0.2597908243017677, 0.30566196392891776, 0.3510286319264555, 0.3918261625254288, 0.41953604369965797, 0.4210514232199934, 0.3821592676656662, 0.2959352023152686, 0.16977165930668947, 0.026180421793775308, -0.10563992463837159, -0.20117379094134427, -0.2514056866099183, -0.26803311943212116, -0.2800852197924094, -0.3201305474728689, -0.40995441595537346, -0.5521579471981963, -0.7295889729829321, -0.9163773033430367, -1.0895771563661893, -1.2332623575450476, -1.340007536712354, -1.4132586262914921, -1.4641745156445303, -1.5005407936608135, -1.5203134977952941, -1.517522427893029, -1.4905906326610268, -1.441413976551365, -1.370681830528381, -1.2776103871149183, -1.164189980464256, -1.0360211985634884, -0.8982870070373679, -0.7528425912134465, -0.600450509553389, -0.4442311716850494, -0.29003000831405484, -0.14226025360330363, -0.030551311309884987, -0.053521349720671615, -0.06058957420889771, -0.046177211736561155, -0.012492294139488862, 0.03184727066843284, 0.07899883520042242, 0.1261146225732374, 0.17478881059538676, 0.22743442415637796, 0.2829831090168173, 0.3343624959972065, 0.3695150871922133, 0.37209114574897056, 0.3254843223877602, 0.22136313848641317, 0.06687190930615551, -0.11299180729372348, -0.2818165118972023, -0.4049773071547929, -0.4664467748070296, -0.4766867513524427, -0.46920529742022277, -0.48502153680222654, -0.5562646483019815, -0.6942760679544783, -0.883222497135726, -1.0897974438432056, -1.2823691936955748, -1.4405807872290883, -1.55645617186353, -1.6336980404641968, -1.6842680383491555, -1.7188563057680217, -1.7382075500098766, -1.7356513744830184, -1.7068772756585753, -1.6525709938502218, -1.5736347389443437, -1.4699868237578884, -1.3439892708485408, -1.2011448523244124, -1.0462066044855836, -0.8805817417933729, -0.7051139852113426, -0.5235896169611233, -0.3427440662021621, -0.16829951696678142, -0.051926733041947004, -0.09824762828898717, -0.12838275766925586, -0.13330212494116508, -0.11413007725058996, -0.07999762152306025, -0.039320905233579534, 0.004444262976216984, 0.053163348837696515, 0.10980107741336868, 0.17104681289680296, 0.22616195266167213, 0.26155052451924843, 0.26071005462493163, 0.2072389265502917, 0.09235817683565777, -0.07851714340919926, -0.2813524212918047, -0.4768196034262253, -0.6239930291272769, -0.7017420293582247, -0.717938831163769, -0.7043193246181239, -0.7026852353754427, -0.7511548360734519, -0.871123516282695, -1.0531503356768517, -1.26118703882225, -1.4568803669820403, -1.6161063061385603, -1.730960357954687, -1.8057810351168813, -1.8518459134105196, -1.880682711582918, -1.8964253252078045, -1.8941512014623418, -1.8669525373760179, -1.8122110818562185, -1.7297963170373583, -1.6201108865103984, -1.486095350996512, -1.3332521161117388, -1.1659249903988507, -0.9854136318762404, -0.7927093487687711, -0.5916298564365791, -0.3889755950294292, -0.1914210859138068, -0.07665430708792392, -0.14793215871426035, -0.20196040228401752, -0.22805397755193105, -0.22639895273894808, -0.2055126506665552, -0.17454953962089903, -0.13813864214186614, -0.09432105775065781, -0.04108486588388265, 0.014361892816288723, 0.059814150580849976, 0.08389278263593275, 0.07338449196225277, 0.014676893538208525, -0.09975387314171677, -0.26595946076391574, -0.46450495216696214, -0.6610119263635738, -0.8168383869032813, -0.9094144168087984, -0.9421668093978288, -0.9388663764721058, -0.9342978521166115, -0.9661152553122968, -1.0638226359522567, -1.2265906466525907, -1.4198167481329125, -1.6022250458024072, -1.7490496384691554, -1.8540794796640845, -1.9220925870964405, -1.96200100332522, -1.9832876150738892, -1.991950973095053, -1.9866401145993953, -1.9606290987401687, -1.9082650405952912, -1.8276572869427832, -1.7190094652235202, -1.5844288226527463, -1.4281073787332141, -1.2537806667798033, -1.0633256342835256, -0.8587680604522868, -0.6439731917982723, -0.42532734430285585, -0.20995925860311, -0.10270349148162103, -0.1981471875980001, -0.2746400761063459, -0.32229426792054744, -0.3401727004884916, -0.33492513861347345, -0.3166165196367504, -0.29195690529181495, -0.26047979425075496, -0.22391959032896708, -0.19234647063791457, -0.17468465206892672, -0.17592785528155638, -0.2036177310147575, -0.26661995945155453, -0.3682812761404332, -0.5036406268403386, -0.6613487638896579, -0.8216658192975521, -0.958307176721995, -1.052166132625231, -1.1017323056579493, -1.1193104134370355, -1.1262465293014257, -1.1523997043047611, -1.2304929079778693, -1.3684720791483092, -1.5367161921478811, -1.6953488980616584, -1.8216139129379834, -1.9122265044276214, -1.972519096685644, -2.0081218021113494, -2.0245162111076986, -2.026394352206188, -2.0150331027914614, -1.986941605395619, -1.9367228920557715, -1.8613720792399462, -1.7603753643535078, -1.6337271898779713, -1.4822511736541513, -1.307957392101286, -1.1133222525959612, -0.9019498886527617, -0.6788488502974519, -0.4502904524173348, -0.22311276355776402, -0.12787090383121896, -0.24536975664757332, -0.3422667325038067, -0.41098526348064085, -0.44897042222463646, -0.46086559171182123, -0.4583689102149047, -0.45163515072334903, -0.44465778407643197, -0.44336857233085913, -0.45579055383111006, -0.48328121169501553, -0.5226169363424118, -0.5753273105746513, -0.6441233858255746, -0.7240863416128214, -0.8056957093353367, -0.8876007575675883, -0.9716173726976829, -1.05244921543149, -1.1205159880580828, -1.171968886261115, -1.2084648094906203, -1.2345235151838196, -1.2650429088533615, -1.328589340180389, -1.4404712614771318, -1.5797452309339655, -1.7118330288197532, -1.8167157903533526, -1.8929142217311425, -1.9465642417647295, -1.9811581281358066, -1.998572984378421, -1.9998031829338438, -1.984590863376149, -1.9524520695363898, -1.90233154097478, -1.8329184630827315, -1.7432764356784378, -1.6312744798360002, -1.4935377759284372, -1.3284870945752016, -1.1378799312244572, -0.926315629496913, -0.7004108505670092, -0.46705363763768376, -0.23255305672023133, -0.1503589345737174, -0.2876622289359342, -0.4030219365638412, -0.4904645020621237, -0.5467398071788583, -0.5770179059523399, -0.5958881914826524, -0.6177066572783072, -0.651005105773888, -0.7015230344288977, -0.770919111816266, -0.8523662779999308, -0.9364443264562299, -1.0197609616688765, -1.096914124619243, -1.1514627918183478, -1.1684385035511993, -1.1551293794366704, -1.1344069041629892, -1.1256796755893042, -1.134407130324271, -1.1601489498630875, -1.199328740548699, -1.2419265810564208, -1.2834314230904713, -1.340174430331979, -1.4300770497918271, -1.5421677917825676, -1.6489630435204876, -1.7341581465693792, -1.7969999727623316, -1.8436489334845307, -1.8775549469505215, -1.8992780800148297, -1.9069248235987164, -1.8955405903301707, -1.8632972928322329, -1.8136972587482456, -1.7503552429243878, -1.6737700474878876, -1.580573190719751, -1.4639809548541658, -1.317928484887601, -1.141298858334812, -0.938074753727387, -0.7153389952349422, -0.48077442878882537, -0.24095028821490339, -0.1688764563397034, -0.32355825021602136, -0.45471174299279354, -0.5568517358824775, -0.6298998389003583, -0.6830901086107591, -0.7332523652193582, -0.796152441341929, -0.8796490935375231, -0.9844414790759455, -1.1062119680345734, -1.2344970805504423, -1.3586663733675246, -1.4701635283021457, -1.5548676367904535, -1.5878478144847208, -1.5507368632118306, -1.4528937990352886, -1.3287335925969792, -1.2169233044236838, -1.1398571117573433, -1.1085948982471514, -1.1235726106974133, -1.1682054561435609, -1.2206269429888015, -1.2798820662852695, -1.3574833195778382, -1.4481765277485845, -1.531518015668307, -1.596631134179598, -1.6445873731973468, -1.680265092701539, -1.7080583511984497, -1.7299768694125008, -1.7440315418890062, -1.7424099118483187, -1.7189378313156298, -1.6768445645412102, -1.623546460693742, -1.5626157233107325, -1.491268456525578, -1.4009401614081614, -1.281594978881776, -1.127959232124893, -0.9414228431242774, -0.72771815773915, -0.4945481756968654, -0.24978045270068902, -0.18296963595698987, -0.3515501410634739, -0.49492930021711057, -0.609690338827475, -0.7026766447457901, -0.7874534778027714, -0.8787218381549053, -0.9870058544791988, -1.113935846040193, -1.2551874561465355, -1.405358817765968, -1.5586746226320813, -1.7067945228648778, -1.834984267580427, -1.925282417892297, -1.953351919181708, -1.8976827979629223, -1.7590658143525797, -1.568800799189683, -1.3713014218640223, -1.202598253198578, -1.0904648756314614, -1.0499209326297516, -1.0704292610105501, -1.1218078045176805, -1.1853332190756514, -1.2575235896987464, -1.331303801691324, -1.3914192672837635, -1.4330195045038874, -1.4616082384262423, -1.4804258983917908, -1.493714573599621, -1.5065820612299794, -1.519549498016933, -1.5261357923665333, -1.5180876547002387, -1.494403957506505, -1.4607082490403667, -1.4214465081339729, -1.3754824610558132, -1.3147002723423333, -1.2266475129947216, -1.1020436889590124, -0.9384156393306089, -0.7384306272163551, -0.5087401116009895, -0.2591304049050145, -0.19275663930992626, -0.37137007707254077, -0.5250880251337035, -0.6551644908307727, -0.7752746694155274, -0.89866557343088, -1.031099768227682, -1.1731139920807852, -1.320534575163797, -1.4681942019984704, -1.6160426230201181, -1.7678562584268471, -1.9181440100429514, -2.0503887129588843, -2.1503117618660044, -2.19951486470967, -2.171804528750572, -2.0522308546935224, -1.855528056789085, -1.6173826256400283, -1.3782625878123829, -1.1799564312035744, -1.0588049846696597, -1.0230900897289625, -1.0475761670869563, -1.100889901813621, -1.1621001185418527, -1.215686879919892, -1.2485165611375675, -1.2599469690684357, -1.2618669201252906, -1.25843792007401, -1.251815919371448, -1.2486745034278066, -1.2527708705146834, -1.2618608279145032, -1.269875589853933, -1.2724681744760078, -1.2692754520626492, -1.2608276209670495, -1.2455973606891617, -1.217007258219762, -1.1622664670139056, -1.06916289335574, -0.9314006940602265, -0.747743253515815, -0.5229407144271689, -0.2687221953892017, -0.19926834403555224, -0.3859379644178559, -0.551553964052742, -0.7025846270121029, -0.8554596676875776, -1.0156718952595165, -1.175889177974996, -1.3289296143678835, -1.4697714926120156, -1.5966737365183286, -1.7175159064442003, -1.8444948706205997, -1.977430132128548, -2.1052999397292016, -2.2215014502455457, -2.314311685002281, -2.3544306092920793, -2.309226791097638, -2.1672710691092916, -1.9442202317849138, -1.6734402796187733, -1.4043066625412808, -1.1943099708838865, -1.0760788427706744, -1.0409187235243442, -1.0558287705625222, -1.0880611780877363, -1.1106705812695545, -1.1078500901006625, -1.0802220317827058, -1.0454429862875998, -1.0147573019174063, -0.9886781371149413, -0.9705706805017714, -0.964723385375725, -0.973224445680699, -0.9947792264084684, -1.0253056791791781, -1.0592737567596353, -1.0902420445385665, -1.1123658138203512, -1.1188623984787835, -1.097790343448969, -1.0355737129852562, -0.9231431199194083, -0.7558987088055569, -0.5364925539725836, -0.2780819721100764, -0.205047962739104, -0.40051171010453046, -0.5818883807421326, -0.7588731778966186, -0.9435936956733243, -1.128771304393089, -1.2963352659327227, -1.4368327229514193, -1.5493521708401095, -1.638364648935161, -1.7178782826475119, -1.8061308109451542, -1.9102555707285513, -2.0284503534151312, -2.163789691190931, -2.3134011222823703, -2.44760010914879, -2.5146582313122336, -2.471876218339491, -2.3071391987845167, -2.040651420689125, -1.7242706520577835, -1.431173549725592, -1.2162471735158071, -1.094091427746149, -1.042507852700673, -1.0262000280585815, -1.0087187111378677, -0.9648239238164907, -0.8927083774654322, -0.8133297761011967, -0.7491420969992595, -0.7052675794541915, -0.6796305977166063, -0.6714448176847684, -0.6830328662179077, -0.7170920070632315, -0.7730641106342767, -0.8445080579283188, -0.9189166037007073, -0.9832905657706938, -1.0273032053878821, -1.0389353638332846, -1.0046940602208203, -0.9144859462024086, -0.7618877453046798, -0.5478647188190304, -0.2862496047815587, -0.21237861709759728, -0.4196810001709396, -0.6209855319132539, -0.8247552108326801, -1.0333709478340056, -1.2271151031891252, -1.3817420545826733, -1.490333857997708, -1.5601655818697338, -1.6036752736402706, -1.6399388290875232, -1.6908283191917273, -1.768257704501938, -1.878670086412361, -2.034758323740184, -2.243288725194854, -2.4743656836435863, -2.660769212086238, -2.7298832150380323, -2.640363626918016, -2.399222992868402, -2.0556139980528094, -1.6893172656279316, -1.373341326616904, -1.1488791443562387, -1.0155055221559819, -0.942043573055566, -0.8865656522163144, -0.8102299810385748, -0.7010456369645984, -0.5798999313043762, -0.47912943405098757, -0.41438901984255405, -0.38424252780583856, -0.3821719234074889, -0.40546578021918905, -0.4555320503165082, -0.5345052471192618, -0.6391543468215021, -0.754995335307948, -0.8618671632422721, -0.9433263027363703, -0.9854607386918237, -0.9751416258896626, -0.9028434240594994, -0.762408130949495, -0.5540467126536719, -0.2914724750551905, -0.22227386641185667, -0.4444629704238842, -0.6673315520945133, -0.8939685004382414, -1.1154497897718827, -1.3029716778796043, -1.4305945486846183, -1.496183444936194, -1.5164345286797036, -1.515032402014974, -1.5184001989646296, -1.5490894916256448, -1.618088934357874, -1.7337658523034467, -1.9129469217488806, -2.1662988422343195, -2.4664362553702515, -2.7427365820670455, -2.9024377051260095, -2.8829131013391716, -2.675180084704038, -2.316203086716108, -1.8834544822926529, -1.465892601819928, -1.1345123628941027, -0.9188608526370008, -0.7967208704019563, -0.7190246242600147, -0.6347796747263789, -0.5142889982609424, -0.36903624314343575, -0.23728769263972146, -0.14720100388441942, -0.1059350433922013, -0.10846855432303985, -0.14746652283056833, -0.2179795951343093, -0.3196724042039266, -0.45193258853473633, -0.6023110033393263, -0.7466277955443835, -0.8617988477235428, -0.9306062717925808, -0.9399818104540373, -0.881876522527911, -0.7518668927469466, -0.5505649153814908, -0.2911921234160476, -0.23333065638351658, -0.47079636157725085, -0.7129304715158953, -0.9554763200663118, -1.17996153111437, -1.3537003402605294, -1.4519918900596784, -1.4757835420182757, -1.4505616472848408, -1.4137316043282473, -1.400676908589048, -1.432803538858029, -1.515822406470132, -1.6536711763031593, -1.8557952664959796, -2.1256137119792107, -2.4443833192355338, -2.7534638597610126, -2.960388055354736, -2.9918885139156024, -2.8159740184967195, -2.447353317152158, -1.9529629379669102, -1.4352494355675958, -1.0021578796856643, -0.7175806866937761, -0.5731871581568799, -0.5047475683096849, -0.4459250457179201, -0.3501142636211385, -0.208505636238988, -0.05595326717805752, 0.06444326650248287, 0.12855828372132888, 0.13234337722183734, 0.08346766760740863, -0.006247606166028772, -0.12919286778928013, -0.2833310515855127, -0.45907491254256494, -0.6318813694432917, -0.773823036140135, -0.864242173722897, -0.8898286539788104, -0.8440208790574578, -0.7246337508583911, -0.533524541204103, -0.2833241843084404, -0.24226305075614732, -0.49165868957260034, -0.7469646927638589, -0.9972454692727307, -1.218928809784452, -1.3801448413984412, -1.458005275668985, -1.4544156176315755, -1.4007880985106187, -1.3448441213759115, -1.328918683184467, -1.3739807422884753, -1.4811281884044993, -1.6449281745663216, -1.8555494196777906, -2.1038861871999703, -2.384050219105584, -2.6630675446924745, -2.869145957621846, -2.925213932559811, -2.7765216974654447, -2.406321963538988, -1.85900890770618, -1.2483992644035493, -0.7280961434523194, -0.4015869073328029, -0.2752640290480686, -0.26374801102100304, -0.2725506501963703, -0.23642110047444828, -0.12483559212612591, 0.03892225572587275, 0.1973360731980896, 0.2999968938496521, 0.32614085842287616, 0.2803476528749685, 0.17956807468566008, 0.04078201351991183, -0.1282179746210433, -0.3195344456023451, -0.5106414968496662, -0.6712799010826496, -0.7781474574552075, -0.8178285531090325, -0.7847017006290006, -0.6783157577228534, -0.5018171566114688, -0.2673793954233863, -0.24663536505057146, -0.5016871237892416, -0.7619612528017528, -1.0125395754928619, -1.2294266681946389, -1.3849463763449683, -1.4582274267803683, -1.449635660449754, -1.3909598880100436, -1.3335366503467045, -1.3218059502969934, -1.376176307652725, -1.4946432574073403, -1.6608526802571304, -1.8468281999589324, -2.0357563769943345, -2.233642701966172, -2.4359314688897786, -2.598601373684703, -2.653797459272418, -2.53213884364889, -2.178527993831151, -1.607813163627223, -0.9372728362270042, -0.35829217706798233, -0.017444941495410524, 0.06210122776890201, -0.01816513372867049, -0.1307629817090205, -0.18328495053723112, -0.12104255547980369, 0.04511263290961239, 0.24476370953519314, 0.39911660849379177, 0.46446887191689845, 0.4382811230900982, 0.3399177353407664, 0.19472427725486038, 0.019416008666643666, -0.17744696230529763, -0.37696806375730024, -0.548837107448498, -0.6679485534904258, -0.7208152838307677, -0.7023102728640048, -0.6127675686487736, -0.4560522203587118, -0.24391207749368984, -0.24538766191389857, -0.4990689776611992, -0.7562303696996902, -1.0011234584025497, -1.2138660525111953, -1.3732179945268947, -1.4593552314398677, -1.46816994720294, -1.4268456518304924, -1.3825112105552766, -1.3745897944995753, -1.4207123471505487, -1.5178446006489061, -1.6459165691101458, -1.7712144465569668, -1.8729531553445429, -1.962764437469049, -2.058622928401023, -2.1469460034707883, -2.18464732794204, -2.0998195766751175, -1.8126072617868219, -1.3019811123444573, -0.6593423465953179, -0.0795693937314349, 0.2587561642181957, 0.3105188409451561, 0.17017791232372015, -0.026031305519230972, -0.16457688432197715, -0.15991663887396435, -0.002871090807533609, 0.22867598996287405, 0.4338483434642827, 0.547476782653946, 0.5554935869940929, 0.47482163479728573, 0.33569230269890865, 0.1638185470412226, -0.029237367520193916, -0.22840417225635193, -0.4052123797790289, -0.5334836767386777, -0.5992904312211215, -0.5976915597082458, -0.5290107912152245, -0.39723933958271096, -0.2135532148875684, -0.23852511490434616, -0.4842102807496379, -0.7313082358395709, -0.966479484341482, -1.1772804736825238, -1.3496637341664508, -1.4640954297895468, -1.5094148612126348, -1.5027640266604945, -1.4800353497419558, -1.4703897173877396, -1.4869282300579199, -1.5290925596116167, -1.5818372724017418, -1.6148103327248131, -1.605887893667882, -1.5676274231921727, -1.534091278291424, -1.5287605887158424, -1.5480889820083539, -1.5360317619003054, -1.396581717615424, -1.062659739090249, -0.5733943544682872, -0.08619002223616806, 0.23159461489911537, 0.31069195424163903, 0.20469571145411306, 0.01558182368829506, -0.15069686500448523, -0.18704703256847788, -0.05141287409155152, 0.18940749382588332, 0.42743705470048954, 0.5852831191935713, 0.635861812828952, 0.5870237132617349, 0.46764259235024025, 0.30915324555456153, 0.1275873776209184, -0.06454755064857491, -0.2417349206364301, -0.37721518633251677, -0.4560764724707375, -0.47324421075785006, -0.42885116393951545, -0.3266842961705442, -0.1770335918147478, -0.22744100644270251, -0.46030263373062624, -0.6926315209206417, -0.9154302466788335, -1.1255583175463602, -1.3171797155040952, -1.470628255733044, -1.5653934005013386, -1.60416747715921, -1.607768931913156, -1.5930040726303836, -1.567760619748607, -1.5346446140060368, -1.4857500798421241, -1.398834448665366, -1.25486187940473, -1.0662161689353742, -0.8845321287394118, -0.7768040649613642, -0.79174504564197, -0.9006238919604664, -0.9800377634025571, -0.9032533268796333, -0.651307381367185, -0.3295955714245607, -0.0692427084984105, 0.059065891098801296, 0.06358982267849955, -0.017678699144180146, -0.12706529213682274, -0.16862923344688052, -0.06182400347936254, 0.16162246523971105, 0.4066827187537844, 0.5952348108109673, 0.689117267622566, 0.6827671340760464, 0.5964874483227325, 0.46133098820373497, 0.2970951086751379, 0.11605795098844411, -0.05872948383047325, -0.20034849589581147, -0.29268514602645557, -0.33020107615828165, -0.3131006861630086, -0.24498668685660543, -0.13478037325365222, -0.2153573581421105, -0.43378538209670553, -0.6489866517615404, -0.856982846758877, -1.06480149009964, -1.2754618087819662, -1.4703822266574047, -1.6203056955801927, -1.7110159448360824, -1.7455198260289895, -1.7282494961296289, -1.6617587488543017, -1.549131127850643, -1.3883836608317286, -1.1675937038416082, -0.8751587318564498, -0.5286194658670803, -0.19609923837538473, 0.015138609975686126, -0.009570682138008966, -0.27494814494630226, -0.6088266107164986, -0.8134178964675215, -0.8167774577688307, -0.6761727111538451, -0.4859046241608777, -0.31249122400511387, -0.18372862103630352, -0.11146055531314752, -0.10132199730991169, -0.11091096926970641, -0.032033839051749384, 0.156041731583224, 0.38839533247121016, 0.5948815734353223, 0.7290012751766484, 0.7707681082439802, 0.7276491424324031, 0.624548149267478, 0.4823025942714103, 0.31472573001509024, 0.1439526204976251, -0.003144965540188124, -0.10928882159274599, -0.16867135530055727, -0.18196450306956669, -0.15241876568497875, -0.08696775875021083, -0.20476852539099916, -0.4095742624348722, -0.607115488067211, -0.7975406631853139, -0.9974892209268771, -1.2198792907957896, -1.45019293513524, -1.6532238334994453, -1.7986230949390674, -1.8703026113810426, -1.8596603368699733, -1.7641023501472088, -1.5861202943668142, -1.3267603452373808, -0.9847982585355614, -0.5612623415227634, -0.07926693527449348, 0.381899794403351, 0.6780417079896411, 0.6356604585518585, 0.22345351081290651, -0.3385348870857828, -0.7930917871390663, -1.022451073870378, -1.0440891006043616, -0.9243344683868929, -0.722950784224275, -0.48866840228791697, -0.26476900172999385, -0.11519816768734127, -0.06492949203631458, 0.003171537757874315, 0.16007628014104858, 0.37731453607215165, 0.5966135523371808, 0.7672672454952909, 0.8580403104301526, 0.8633383952275803, 0.7980653331166785, 0.681084325919085, 0.5285136332531772, 0.3631692240393872, 0.21163355591405675, 0.09161080519756083, 0.008863059453249639, -0.03770461331551645, -0.05070459690506931, -0.03449280490365631, -0.1956410284483593, -0.3879609240993524, -0.5679224815097921, -0.7379492604703929, -0.9224845274973642, -1.1445227892890917, -1.3967521034378627, -1.6433142134833882, -1.8423232736616455, -1.9594999819933085, -1.9718684476672144, -1.8702908513899397, -1.6561409746694058, -1.3342283927143097, -0.9150791626766337, -0.41167757666859683, 0.1472931401618487, 0.673192745040747, 1.00332799358495, 0.9513573652314217, 0.46573585967099396, -0.2256348433992998, -0.8433125492994816, -1.2368329540395289, -1.38810967778557, -1.3335082680826993, -1.124574112372712, -0.8210432283369375, -0.48875111181025704, -0.22653243612710167, -0.09944054548813545, -0.0037306999974490948, 0.15314300221791827, 0.37060053628538964, 0.6059821202575687, 0.8111777992592305, 0.948502831651037, 1.0019396581397637, 0.9760558013948782, 0.8859036719275221, 0.7497062185395879, 0.5916660026687811, 0.4374681446808427, 0.3040543752010966, 0.1968219024900634, 0.11467978512586598, 0.056430634373944466, 0.02071800116566999, -0.18613227719983452, -0.36582090102681797, -0.5282301830997937, -0.6759680068937592, -0.8379124715864267, -1.0463711295308142, -1.3039219130555608, -1.5793082481646747, -1.8256289506817727, -1.9951605648969513, -2.050171129212809, -1.9730030881169722, -1.7643937632517508, -1.4333323989527706, -0.9999181695723941, -0.4879164376618872, 0.069610787681588, 0.5835487666944573, 0.899342008868032, 0.8526609970390425, 0.3990347181614421, -0.2903197837697481, -0.953750844279336, -1.4263694772783857, -1.658662050637323, -1.6551000711005759, -1.4582170180093763, -1.135027719844024, -0.7622160672294829, -0.4394088471050424, -0.23221675565967154, -0.06974471577834078, 0.1270220267673759, 0.36820083189338704, 0.6269099212231025, 0.8643190792224398, 1.0424956402857901, 1.1388467241761084, 1.1495871876069608, 1.085780056846331, 0.9672775828079103, 0.8190083863517231, 0.6644319092479246, 0.5185622749034037, 0.3863552631664639, 0.2675570324992947, 0.16346015136556438, 0.07579360619442303, -0.17435949119470784, -0.33982681607899906, -0.484365498736198, -0.6095249060067899, -0.7449162609108346, -0.929165206897269, -1.1756008403577354, -1.46223371636049, -1.7442133094032704, -1.9673990910486938, -2.0821973295578613, -2.0606344815412743, -1.9005552012367388, -1.6145672975549903, -1.2292286142088515, -0.7722440271421237, -0.2750067969353551, 0.17886150736864392, 0.45796456550514114, 0.43685466715763405, 0.0799510074205744, -0.4988067447861335, -1.0941935619708165, -1.5545726340787704, -1.8065437228951435, -1.8303261847463996, -1.6612290087393562, -1.3646236095860491, -1.0090750673105633, -0.6713082309864233, -0.3991551792873078, -0.15738493752114244, 0.09961371577763371, 0.37755628407590813, 0.6631588217807686, 0.9284669052607191, 1.1381566984800306, 1.2673979796093722, 1.3079309920744289, 1.2681126200322486, 1.168254574236889, 1.0320002367622856, 0.879146410532755, 0.7222023170390265, 0.5658323968236626, 0.41156723206463935, 0.2639145527926998, 0.1274673862681757, -0.15944146102737894, -0.30857663298444243, -0.43544817705621874, -0.5399282378313638, -0.648656537014066, -0.8024617822545622, -1.0241282801503608, -1.3033829723738395, -1.6037090934795075, -1.872426937344031, -2.053780849710188, -2.1100832061443477, -2.0339351848868583, -1.8368783578071723, -1.5442322686097623, -1.1841550316883496, -0.7822713821512688, -0.40627568371890826, -0.16443920574637927, -0.1469263329407653, -0.38081832380770875, -0.7946394470622564, -1.2446972710286215, -1.6083476637852077, -1.8136313854501738, -1.8358721835325598, -1.699860026886665, -1.4558823639180476, -1.1493152767181076, -0.8257716500327009, -0.5156782119891281, -0.2125640338971603, 0.09684330472590019, 0.4098055948423441, 0.7187173266810363, 1.0017327485155338, 1.2289262074126823, 1.3780322027396597, 1.4394660255299914, 1.4200608367009961, 1.3388491054159541, 1.2159213529623287, 1.0664145600436659, 0.9003500977860476, 0.7226045414141719, 0.5370870521123153, 0.3514982472123411, 0.17266629140807047, -0.14209379037808803, -0.2737998275209016, -0.38417261225627464, -0.4714648782105017, -0.5567979785076869, -0.6782537121096182, -0.8649354605494818, -1.119063999542439, -1.4165760127233684, -1.713448813015682, -1.9553132723111581, -2.0971336257178863, -2.1226319360086983, -2.0377136117469528, -1.8605910718418546, -1.6154141402703526, -1.325435880922716, -1.039116727748548, -0.8355576611922593, -0.7802426763521922, -0.8922846150252326, -1.1298946564870698, -1.397255851262067, -1.6054801755876458, -1.7095945434682986, -1.6996129993254654, -1.5889109557017747, -1.4016447323187595, -1.1525302002116, -0.8571052190292898, -0.5358605848749362, -0.20070476881384752, 0.1387457151391514, 0.47135705956918933, 0.7888734057837768, 1.0735779779096999, 1.302910194085181, 1.458897093232588, 1.5325504754610044, 1.529760018755135, 1.4662594707831222, 1.3567046268473715, 1.2115561384124454, 1.039115762919931, 0.8449032097941492, 0.6351702156408768, 0.4202896644467389, 0.208388868356047, -0.1238863961519179, -0.2387827261319979, -0.33497089146203796, -0.4088236874500408, -0.47510208764395445, -0.566217299488166, -0.7133073180709881, -0.9288253773413506, -1.2025257289753306, -1.5040448336485406, -1.7872191440958847, -2.0048651613903763, -2.131245619486144, -2.161997628262919, -2.1041279069495356, -1.9756285083009815, -1.7976773780122421, -1.6052308855491815, -1.4493165076433558, -1.3708790022187924, -1.383078054644027, -1.462119174017231, -1.5492231755256676, -1.5882416158459445, -1.5676325738941892, -1.498613061122036, -1.3884045242429504, -1.2377345529776254, -1.0340091342134514, -0.768057115724851, -0.45394394245585773, -0.11536612247245297, 0.22437440625436944, 0.5511985541348513, 0.8561676566742153, 1.1255966371803645, 1.3438552437420197, 1.4968448977120927, 1.576664841163925, 1.5874102820958733, 1.539738615264872, 1.4423499504512196, 1.3021812360145222, 1.1266595977154394, 0.9221905260536993, 0.6972980402899832, 0.46419678403894316, 0.2313582218075804, -0.10619477690158388, -0.20626186917149064, -0.2916648876466803, -0.35618362277562665, -0.4080156924351148, -0.47288390569466365, -0.5809316145485756, -0.7512864197359643, -0.9852980344496521, -1.2676470634466475, -1.565924400007084, -1.8357674433968934, -2.0429582817123926, -2.1740369352423823, -2.225616843167936, -2.2059219341698797, -2.131943760232113, -2.027432675967157, -1.9249432400484612, -1.8449230282775766, -1.7927012365715131, -1.7556187462106627, -1.7015079218251232, -1.6012359189718555, -1.4664304434294928, -1.3234078303333738, -1.182121263405509, -1.031744697539152, -0.8436272678188074, -0.5965221908069226, -0.30108906516437794, 0.013950716040771808, 0.32475037975551124, 0.6208092257532425, 0.8946968852441133, 1.1367052095499943, 1.3360514207123677, 1.4810499274673312, 1.5639695307699237, 1.5859099309827798, 1.551576157227933, 1.4646101063003751, 1.3300324395469796, 1.1551373374993548, 0.9474797436614913, 0.717685091277312, 0.47887716284821, 0.23921239283014786, -0.09014263591857825, -0.1783125104526289, -0.25705829724800844, -0.3168929475708859, -0.3595371004901225, -0.4039267381323584, -0.477254399850879, -0.602090950816039, -0.7880550458116988, -1.031799513044159, -1.3167731880549518, -1.6088623913889566, -1.8688244843936093, -2.0729216929472325, -2.2113533724760654, -2.283254487398584, -2.298395026480482, -2.27189632121194, -2.22331252936862, -2.162385639246807, -2.086084420866669, -1.9877388163608558, -1.8505959870640152, -1.6633441422923523, -1.4463231654187951, -1.2333408632353384, -1.0399954588125198, -0.8556162121946799, -0.6523237189804769, -0.4115336310853851, -0.14259784082776544, 0.130724402098716, 0.3952621176848671, 0.6473554808943863, 0.8820930677217381, 1.0926039089724788, 1.2709059886801686, 1.4065374074944899, 1.4911534124529515, 1.5220921961294476, 1.4983322638263257, 1.420454896011227, 1.293022336394828, 1.123464104404958, 0.9205868404052103, 0.6967209103525256, 0.46489625658759615, 0.23229912356065577, -0.07749657538765066, -0.1565925490332617, -0.23179779970338749, -0.2916461073351968, -0.33151536296083545, -0.36286171311469206, -0.4086564171180408, -0.49275185238223157, -0.6302120988438527, -0.8251958379894099, -1.0729016346381104, -1.3539648094089516, -1.6344795744459528, -1.8825695640748554, -2.08056180502074, -2.2212435035662943, -2.3062601530434854, -2.3446368573334087, -2.3477417094172925, -2.3197384974607105, -2.25287894245496, -2.1409302468916462, -1.9757524512530615, -1.7540675387090565, -1.4957086502836603, -1.2345154013142905, -0.9894221988750943, -0.7585225860480829, -0.526791050072773, -0.28617112776450737, -0.04552988086441057, 0.18386430202756493, 0.4023545250602149, 0.6117947500370142, 0.8094351231907332, 0.9906099253678488, 1.1490220303167271, 1.2750440330691015, 1.3597031284425158, 1.3965914736590206, 1.380873926415602, 1.3121353697662486, 1.194893664694955, 1.0365791762810705, 0.8472898120848896, 0.6402672443259979, 0.42713303634047234, 0.21340359250202082, -0.07001147791874592, -0.1426243780027627, -0.21517175048710765, -0.27755563063314886, -0.32092123181266585, -0.348664264102509, -0.37701571152961433, -0.42878763120195146, -0.523154392251909, -0.6689263226804548, -0.867245689442098, -1.1105513488966305, -1.3778390797945486, -1.6392151285950107, -1.8699258378556856, -2.0561308473471773, -2.1922930785628028, -2.2811892172993447, -2.329565115128555, -2.3385796983189, -2.299209086497441, -2.203607293762411, -2.048164359766686, -1.8323663000073755, -1.5709119927025754, -1.2912274783295683, -1.0141767439041873, -0.7487734063477328, -0.4957069624617655, -0.25832994197173603, -0.042596468512072605, 0.15361494169556528, 0.33802162690395393, 0.5144752345846069, 0.6820623531483493, 0.8382881972999425, 0.9782797996452186, 1.0934909453882855, 1.17496391991308, 1.2141836559532622, 1.2050662188915398, 1.1469797295303221, 1.0440453749391672, 0.903666702439482, 0.7369512010113191, 0.5566502248586894, 0.37173152810367177, 0.18587127765851022, -0.06863254912753095, -0.13737541644211046, -0.20643661788153275, -0.2710672239770903, -0.3216361754030171, -0.35402792658963333, -0.37655923052237894, -0.4080222004581453, -0.46900754663603356, -0.5717297802304369, -0.7199738133690857, -0.9125285417689919, -1.140696076934719, -1.385148826642587, -1.6214008838599605, -1.830361721144688, -1.9998417228752352, -2.125530938816909, -2.2094250772466615, -2.251176400216969, -2.242328679100771, -2.1746964029562914, -2.046333306745789, -1.8581678063245826, -1.6189904215307598, -1.3484431090476203, -1.0684991037372829, -0.7962569893958011, -0.5439917898065935, -0.32001120772891256, -0.12400940769228101, 0.05224327609527117, 0.21586599502666612, 0.36981315633764017, 0.5142764102026438, 0.6489516968765172, 0.7704204061385623, 0.8721491495453673, 0.9465810525066123, 0.9853906274451861, 0.9828182601685128, 0.9379790281540454, 0.8542417539191833, 0.738668182905553, 0.6022687521005864, 0.4559204313096065, 0.30548940674186914, 0.15312303444478992, -0.07245319282513138, -0.14009415482246804, -0.20501165952338915, -0.2693997738664928, -0.3269140126249473, -0.36866634599967374, -0.3951638888228073, -0.41912719088676953, -0.4591665336339263, -0.5292426627010062, -0.6346489317105075, -0.7765375406250089, -0.9538888942279767, -1.1592866681421605, -1.375720312852829, -1.583704540497364, -1.7666168677387646, -1.9133604817492853, -2.020039777462398, -2.0843444535257656, -2.099496728152742, -2.058165462698516, -1.9591622521185121, -1.8048027705055925, -1.6009340061536923, -1.3618988160345926, -1.1082780605758271, -0.8600670724174796, -0.6322231799167998, -0.43072083642154463, -0.2516312239440153, -0.0886542652419793, 0.0610664635123387, 0.19796955903850993, 0.322776277911773, 0.43677357977465114, 0.5383890309806686, 0.6243443192068747, 0.6893499222353828, 0.7267561574670541, 0.7317659492924513, 0.703066453520475, 0.6430739126053728, 0.558062548144934, 0.4574040569713124, 0.3488499142999504, 0.2355137650623722, 0.11864793209016192, -0.07903706507034193, -0.14844875489522988, -0.2100611439108135, -0.27103073682829565, -0.3311872044692832, -0.3823674463394434, -0.41907711983306767, -0.4462827948700003, -0.47755482205402416, -0.5264951282747918, -0.6000493683425429, -0.7005835510633618, -0.8298754453753906, -0.9882745398353322, -1.1682799769931356, -1.3544597381726788, -1.5297451155094508, -1.6790122338025357, -1.7930409849210325, -1.8666475553894097, -1.8947770872089411, -1.8719762011446934, -1.7981788168035602, -1.676867123169115, -1.5139341799955122, -1.320420490660362, -1.112909814172093, -0.9083769904089296, -0.7179685823426512, -0.5443414399817683, -0.38435387835932, -0.2362567236470763, -0.10137268213139573, 0.018244988483747136, 0.12323411333614258, 0.2158367241469959, 0.2971466962833009, 0.3670714435274229, 0.42259436447960347, 0.4592583947428017, 0.4732535398334324, 0.4628943358876979, 0.42974763266923494, 0.3785443759128101, 0.3156580030482871, 0.24519731231077313, 0.16826702375488123, 0.08575950377321802, -0.08575500093768451, -0.1597351136992702, -0.22063089688955806, -0.27630345917031623, -0.33300084264486524, -0.38846863840213336, -0.4360681728237972, -0.4733636218358292, -0.5060398976415234, -0.544880464140251, -0.5982510132032738, -0.6700726943624177, -0.7630958474931261, -0.8802988875643749, -1.0205027585003907, -1.174016191483081, -1.326511408958552, -1.4627557440563692, -1.5702109386266718, -1.641160849031484, -1.6715158751935153, -1.6585491557618162, -1.6034319391318441, -1.5108336096798984, -1.3871118807400926, -1.2406953720567637, -1.0833193044295257, -0.9261792605899825, -0.7759036825039025, -0.6333395263929554, -0.4975022896178668, -0.369987846724544, -0.25460330684802246, -0.1550702595996567, -0.07112225532214156, 0.0002331454983911925, 0.0627515934851468, 0.11841109014382818, 0.16630911991965475, 0.20420429351262495, 0.2291487046801644, 0.23901644584608, 0.2339894699977737, 0.2163012485578831, 0.18903951388416865, 0.15346170526837366, 0.10933240871036093, 0.05722625391040994, -0.09127828429995742, -0.17157761516627118, -0.23523763206843126, -0.2864767098648518, -0.3352502212545553, -0.38710166986454686, -0.43951503506407746, -0.48709216690314394, -0.5279408915242668, -0.5669179463686733, -0.6115880853605589, -0.6672426529072458, -0.7374302889040284, -0.8254356988525537, -0.9324089130335453, -1.0533707591688477, -1.1777459745079382, -1.2929046309136973, -1.385716766023969, -1.446871689704236, -1.4725781740153248, -1.4625061556072938, -1.4189085712247167, -1.3474846802121256, -1.2544816638533247, -1.1466652207954133, -1.0315814185830945, -0.9160132156494206, -0.803112400874596, -0.6925769678206964, -0.5847736096140417, -0.48251592263492843, -0.39061715521300727, -0.31320469434672044, -0.25023911173602037, -0.19806847165715727, -0.1511532896238593, -0.1062491524896067, -0.06228348605205185, -0.019868811760059807, 0.01838328473471521, 0.04938522530876077, 0.07111899001822396, 0.08327740794350928, 0.08637155088642873, 0.0798932266314206, 0.06264083875298937, 0.03480566571323327, -0.095629444172119, -0.18226275900760058, -0.25124305051965345, -0.3013455528354555, -0.341779225353574, -0.3834055310050169, -0.4305988061780783, -0.4811493211376391, -0.5302284447375174, -0.5762791585759666, -0.6225759761634614, -0.6734645275980866, -0.7324273311197101, -0.8027112562836177, -0.8859593598754242, -0.9797866630004516, -1.0774226840813552, -1.1693854376265769, -1.2444786523386997, -1.2932062790011947, -1.3119204622844922, -1.3010715074220633, -1.2636808789768872, -1.2057120922920022, -1.1336110529696923, -1.0531977827608519, -0.9696484878250858, -0.8871489783196783, -0.806338607469417, -0.7259909020718951, -0.646592924502461, -0.5712798199008197, -0.5045589820182033, -0.4495861871495097, -0.40610896184638995, -0.36949793054832375, -0.333461443605513, -0.2940607225447612, -0.24912299999951498, -0.198932922213684, -0.14656573635626666, -0.09631149814675441, -0.05194699617856187, -0.015659821807532132, 0.011163525321396469, 0.02685937470763746, 0.029482409891146533, 0.01899036719449954, -0.09924890460332919, -0.19023211114813707, -0.26384658013825013, -0.3155283372573888, -0.3505539401386064, -0.3801386906071416, -0.41423564649860556, -0.45733949367585236, -0.5073248778278606, -0.5599229207444056, -0.6131118671974278, -0.6670778454519909, -0.7234676846429671, -0.7846488875748703, -0.8521769289058349, -0.9251455812974558, -0.9999187521166552, -1.0698543045637277, -1.1266225281362163, -1.1624951203094689, -1.1739505510955575, -1.1611698251743827, -1.1271956151215028, -1.0778375735843444, -1.0197495975509134, -0.9582032380994091, -0.8976053280757946, -0.8403086660083745, -0.7856920607882115, -0.7319446067692834, -0.6794457715927963, -0.6309536974070916, -0.5892724207886412, -0.5557036110980006, -0.5287979079134097, -0.5031364566224952, -0.47286859851191104, -0.4341078073922332, -0.3847665778429082, -0.3258358000937118, -0.2613101629753686, -0.19615597075508534, -0.13473322315619998, -0.08084094337725567, -0.03737843935292618, -0.006680544175583249, 0.008713094175260332, 0.008999181395346416, -0.1012154542001495, -0.19272571799172078, -0.2666051263433414, -0.3185749419132547, -0.35064168822952146, -0.37119573467914524, -0.39148253726758175, -0.42114506724499784, -0.4634661641655174, -0.515575170718901, -0.572690758221462, -0.6304705460282024, -0.6870643914957406, -0.7430941102452252, -0.7995738886877699, -0.8566054475869208, -0.91253091554412, -0.9633783719572467, -1.003045376625008, -1.0264159436107774, -1.0306751116743573, -1.0158340167458155, -0.9843425857563204, -0.9417789755524006, -0.8944112352470213, -0.8471786415320943, -0.8038922781240557, -0.7661085998869839, -0.732889001176604, -0.7024041452270473, -0.6749148387512043, -0.6517135527771266, -0.6329770406990161, -0.6177365406279185, -0.6031080538650877, -0.584184369637198, -0.5564552488167772, -0.5165951784930237, -0.46313068429912313, -0.39816389635023486, -0.32622082248165757, -0.25179271728898645, -0.1794434318720541, -0.11487948693793336, -0.06224512448030075, -0.02395687769408332, -0.002426731094645517, 0.0033208878061386227, -0.0986444415844289, -0.18536075920344144, -0.2534153425893812, -0.3008434431592139, -0.3292308649706733, -0.34433483719727453, -0.35566614272845976, -0.3737905273735178, -0.4054539192574992, -0.4504000312099298, -0.5037908257518092, -0.5592678703883548, -0.6122048940421717, -0.6614787178993511, -0.7073794635864683, -0.7504400755364776, -0.7903968549720788, -0.8250305729841014, -0.8500928140728525, -0.8624826423585655, -0.8607156328259828, -0.8446451392229118, -0.8164743633537972, -0.7810690969828419, -0.7437492935297307, -0.7089095371851023, -0.6797846415896346, -0.657773820496731, -0.6419989071126905, -0.6310408648212533, -0.624630148030455, -0.6221773015563091, -0.6214061161791008, -0.6194074115247188, -0.6127593893193934, -0.5979000149339647, -0.5718580573510066, -0.5322680110446092, -0.4786816712683156, -0.4138247457453339, -0.34147130090540595, -0.26489516172834743, -0.18925392775417696, -0.12190180337442884, -0.06777767859357176, -0.028927124234408193, -0.006747140409056985, 0.0006593671281814491, -0.08786461894933888, -0.16317319040671857, -0.21985628550654984, -0.25790571311877847, -0.28009396486761096, -0.29093066329646927, -0.2978448639906415, -0.3096377814062559, -0.3327655122532963, -0.3681372762991888, -0.41172022062373836, -0.4574798815424181, -0.5005668954769773, -0.5390148912788721, -0.5727004448510801, -0.6021797278560671, -0.6279925141304015, -0.6489100861161337, -0.6624716724258377, -0.6667697228524245, -0.6614427501509437, -0.6463242347022874, -0.6232925706149676, -0.596068989000593, -0.5688331558339222, -0.5450884962119831, -0.5276784773605627, -0.5178168388225968, -0.5148266088073593, -0.5176066420542311, -0.5252067685099652, -0.5356986236460048, -0.5455668493021208, -0.551063345173219, -0.5489356517829017, -0.5368754246808088, -0.5133969419562358, -0.4775546075752635, -0.42984800188061295, -0.37244667051787866, -0.30770954481048196, -0.23801012096954882, -0.1686533968237703, -0.10736763906801347, -0.05915832473922316, -0.025408908186634462, -0.0063680482774628, 0.0001776179744554502, -0.06671232259245641, -0.12310747742995932, -0.16403577738672942, -0.1900783317154115, -0.2045216066793124, -0.21116376999742512, -0.21535996048125125, -0.2235479123164522, -0.24011787981475244, -0.2653375116557438, -0.29601820144215196, -0.32768840862887416, -0.3569821337293098, -0.38224096966134435, -0.4032810524983431, -0.4205822265114312, -0.43472193336272036, -0.44522145278790953, -0.45083955067124437, -0.45069289809067964, -0.44479948826236326, -0.43312695771559145, -0.41678269188739897, -0.39831836098408574, -0.3806267811127445, -0.3662309403499316, -0.3574089520269945, -0.3551355158391847, -0.3589127522945865, -0.3677115567991183, -0.38033713670942065, -0.3945276097723777, -0.4069725853514754, -0.41437540578939086, -0.4141773910378887, -0.4050189514404575, -0.3866039834566813, -0.35916379761915096, -0.3232660784975869, -0.27999663469009745, -0.23071496312912096, -0.17740095281784132, -0.12437193667612391, -0.07786585404871993, -0.04193795866264828, -0.01733135744816064, -0.003699109265987947, 0.0006926937179713032, -0.03617902303062271, -0.06654946255696048, -0.08815189196341962, -0.10135075712178497, -0.10820747684806088, -0.11114668554646207, -0.11308988181382736, -0.1175512873695136, -0.12668901740974586, -0.1401866444879994, -0.1561475029750622, -0.1721641525407308, -0.18664209459238576, -0.19887701290074566, -0.20868974366963516, -0.21637251412903613, -0.22224027400457821, -0.22619674048776672, -0.2276171331209787, -0.2264694539464854, -0.22269544413717313, -0.21630581470167864, -0.20778460454888267, -0.1984292950231127, -0.18963449323761458, -0.18289355851519387, -0.17949501641523516, -0.17996713341362747, -0.18404061366424676, -0.1910353655184719, -0.20007031978707407, -0.2096267638432922, -0.21778431062698053, -0.2226756700549366, -0.22284788990066198, -0.21771596491707831, -0.2075009242923673, -0.1926408458913208, -0.17326336214966273, -0.14978032447571232, -0.12290925562278224, -0.09394965176825294, -0.06529971482419668, -0.04031479762194776, -0.021216092870732776, -0.008319386481941752, -0.0012687124337201143, 0.0007960894439121046, 5.44868955656516, 0.04734979794646056, 0.08400723853386556, 0.11258378604048287, 0.13744163924131425, 0.16127581506832347, 0.1801811379805804, 0.1921032890450011, 0.20066674836803966, 0.21203770128135316, 0.2269880970702928, 0.24020590493494112, 0.24745764788567537, 0.2523752604586315, 0.2600532556907248, 0.2732105980036449, 0.2897356661175578, 0.30428312079156533, 0.31317574425344086, 0.31474239031670215, 0.3116908338383549, 0.308708308692218, 0.30148872463646587, 0.28453446089802703, 0.2557804485015713, 0.21997443740804484, 0.18049161184522564, 0.13593540257231504, 0.08385406606349106, 0.02487408057075069, -0.031068228498639126, -0.0799336845422008, -0.1261919262647385, -0.16734594165507072, -0.19938251037740923, -0.21362803232254174, -0.21317056559182426, -0.20881346851245589, -0.20356800029690297, -0.19528985738002366, -0.18472079308806263, -0.17518457088812264, -0.169792754941379, -0.17193948414468035, -0.1773654488557515, -0.17527365396833158, -0.15704915642158193, -0.1211420100254873, -0.06716788425361586, 0.09085718464864447, 0.16405506131602715, 0.22413622849871165, 0.27605749698825405, 0.3228549252450851, 0.35918711721921925, 0.3819069740190841, 0.40104109239907654, 0.4253092798959953, 0.4507452150069425, 0.4706014472674195, 0.48062334062953777, 0.4874954938251725, 0.5001059518431256, 0.5205947604671513, 0.5473626066462737, 0.573648024734798, 0.5920460145194694, 0.5964976758590538, 0.5899240286890532, 0.5788735814471291, 0.5614272233705229, 0.5285864107512028, 0.47543571733146617, 0.4074289185566199, 0.3307400125294982, 0.24393094686575523, 0.1431411538253276, 0.03150254194614655, -0.07242044339635113, -0.16058372245816313, -0.24064750222739528, -0.31049347087697265, -0.36549422698616757, -0.3915006369561937, -0.39316506222000536, -0.38639846422092156, -0.37669118379029637, -0.36203313187436437, -0.34329163283412345, -0.3276536080383145, -0.3217419571896048, -0.32604409369538834, -0.33217163709776304, -0.3235873438204188, -0.28383583620666863, -0.21311018624637748, -0.11616446004005994, 0.12496045449154738, 0.23125181385559174, 0.3209654121642398, 0.39921755073600423, 0.46687527401141304, 0.5180380557538221, 0.5538107912707402, 0.5873741668122157, 0.6228786740679872, 0.6531770445706171, 0.6731814577047461, 0.6809613918080026, 0.6868367109381022, 0.701457660297811, 0.7227942198987579, 0.750771980922883, 0.7839863760056122, 0.8129069627154722, 0.8255732314069452, 0.8196808282664979, 0.7983848389210193, 0.7648413368696011, 0.7120708926844157, 0.6348861760824472, 0.5378301673678932, 0.4296424822900886, 0.3060795904715408, 0.16303209055236845, 0.0075313504033574915, -0.1353825043174518, -0.25180015692019503, -0.3486824574716578, -0.42935195315039526, -0.49042825251013017, -0.518610198283944, -0.5184716725387417, -0.5082244930875474, -0.49397583229825254, -0.4732970220429246, -0.44942943905558835, -0.43424773903236596, -0.4357792028056925, -0.4472345836191734, -0.45218520715161786, -0.4323527959696036, -0.3694546629523583, -0.2694408842804928, -0.14400256663307787, 0.14300907303426835, 0.2726934011445376, 0.3859359212186874, 0.48558640604526, 0.5714395301959277, 0.6391635939146041, 0.6935275611991562, 0.7444583798582717, 0.7892285007828469, 0.8192759905962103, 0.832601505576844, 0.8345402815240469, 0.8405755800267121, 0.8580337441849573, 0.8771391916734689, 0.9000406958579293, 0.9351774023491711, 0.9718397145005224, 0.992582390973096, 0.9888663939134181, 0.9588478368909747, 0.908300686915924, 0.8328582803029297, 0.7287750871946502, 0.6019017493558656, 0.4638899720829138, 0.3055502063763769, 0.12483970723432464, -0.06579349856689615, -0.23708232670295332, -0.3712110672580901, -0.47129851386005106, -0.5455071422336527, -0.5931309343062997, -0.6085451254483337, -0.5953829697518529, -0.5730096593543048, -0.5507621349716276, -0.5246269481904094, -0.4986560181122788, -0.49028699268058445, -0.5061851869897899, -0.527875660959446, -0.5296732124255116, -0.49605028432007203, -0.41309498565849334, -0.2938766922834425, -0.1546734125432943, 0.14657831594378387, 0.28410211235732613, 0.40949811529710173, 0.5245183869576476, 0.6276609925450747, 0.7142739124822045, 0.787171360772709, 0.8531618002214337, 0.9060455231841504, 0.9360933541114798, 0.9446845096821598, 0.9441010412098371, 0.9520983971364132, 0.9705584241959497, 0.9831368657762748, 0.99449515275256, 1.022587079790517, 1.0588124946048232, 1.0813450121315649, 1.076654356256183, 1.0378404110582695, 0.9698970347468144, 0.870541356044374, 0.7387835749046364, 0.5836216803772989, 0.4170046848554241, 0.22799520119925118, 0.017122916424215425, -0.19805264176745882, -0.38455610248380573, -0.5237308652564536, -0.617657669689772, -0.6770387506510598, -0.7003385047407742, -0.6886162334029358, -0.6497764063615111, -0.6045433443760243, -0.5664253017294837, -0.5340996135288514, -0.5102453688716708, -0.5101386130079492, -0.5359646375377476, -0.5622397712492786, -0.5586652520870387, -0.5120655902521595, -0.4159298472142179, -0.28944041522072383, -0.14997998035151303, 0.13947252325775727, 0.2736155362973164, 0.40005007917514734, 0.5239978907525051, 0.6437032614819366, 0.7483603884711777, 0.8341145091415176, 0.908043755246173, 0.9659745732619316, 0.9998655765197896, 1.011692004112403, 1.015706845031097, 1.026958159155006, 1.0405797611744165, 1.0385528054854183, 1.0282263254617505, 1.0341666828794498, 1.056773616745351, 1.0692643565531361, 1.0552086645984597, 1.0042958532810182, 0.9205921619681603, 0.8020343257254937, 0.6483173098147192, 0.47004287904198194, 0.27978137230396183, 0.07071883379250322, -0.15524798674260532, -0.3773284448384201, -0.5640414411082297, -0.6989737599893477, -0.7845783936479891, -0.8288078665720495, -0.8266816420826655, -0.781007686477026, -0.7064936952261053, -0.629050881327847, -0.569232310492939, -0.5283608164746204, -0.5065063263726157, -0.5108356068401995, -0.5365376426725298, -0.5563863201651403, -0.5418938527576109, -0.4838839684373964, -0.3841387114588648, -0.2613304425846643, -0.13133310243081303, 0.12396408566722408, 0.2462293048850238, 0.3659979247612455, 0.49309501688996343, 0.6248937254946402, 0.7439976581700782, 0.8396152032949108, 0.9189806986719716, 0.98043462425194, 1.0193085732276552, 1.038652695454915, 1.0520246688487238, 1.0654362434858524, 1.0672942305009763, 1.0442994593269963, 1.0054329319070352, 0.9782621682815275, 0.972800200328874, 0.9616669366806, 0.9274598244793493, 0.8600511647938991, 0.7614092212238456, 0.6291721667771645, 0.4604691051710262, 0.26353937519824394, 0.053702514090566686, -0.16505554023033528, -0.3902673244911392, -0.6023593029344367, -0.7744540170872082, -0.8924323059097838, -0.9618042337057285, -0.98779983576365, -0.960486774446763, -0.8821126039016434, -0.7726279511998408, -0.6633948870999786, -0.5799525787061673, -0.5260730101281, -0.5007545629806472, -0.5030966116368891, -0.5182586356311006, -0.5204671005393782, -0.4904623256048832, -0.42436259123348724, -0.327840071466261, -0.21747388820156088, -0.10593699786223648, 0.0985852707526157, 0.1993508076380499, 0.30674724539061243, 0.43324999219676397, 0.5732538014168935, 0.7040342500201144, 0.8071791857301006, 0.8900221069711406, 0.9547820294576559, 1.0006784530775248, 1.031125090440566, 1.0535389364695413, 1.0645111545929091, 1.0538999934374185, 1.012615542240801, 0.9492813871410257, 0.8892025793525311, 0.8447506157595563, 0.7941213873553666, 0.7269265479336801, 0.6378707498503267, 0.5242128330691779, 0.3779082478729895, 0.19365947638649092, -0.020104881622068714, -0.2433353333740449, -0.4648054452913531, -0.6807334895780227, -0.8717739813230498, -1.0163512814643763, -1.1036614987367355, -1.1420465108502538, -1.1371423684674096, -1.0806904542530305, -0.9722587374176205, -0.8329592016919053, -0.6967220459077328, -0.5900897756233829, -0.5199127371893608, -0.4850195639880322, -0.4795688251579805, -0.48261235670393227, -0.46884029440722197, -0.4261280206314861, -0.35529324233505416, -0.2637253018649513, -0.16711819847424722, -0.07787546265253247, 0.0668688771643293, 0.13878561564525865, 0.22738590531067168, 0.3502508191883812, 0.49845732597477593, 0.6401039906570598, 0.749259925623849, 0.8327822853074814, 0.8993315406313248, 0.9521861893765552, 0.9924096965568281, 1.0197503162587478, 1.0261504685562903, 1.0073258745373643, 0.9576399238011091, 0.882379199136203, 0.7972831517552152, 0.7110047175908029, 0.6129793513082532, 0.5056847085898513, 0.39010780733586825, 0.2561673043720638, 0.09033719185381753, -0.11454345807528733, -0.34597906986017957, -0.5779359994233052, -0.7980436633094651, -1.0022611759983924, -1.1687060036683616, -1.2767828799145318, -1.3209198935225495, -1.3132416456257066, -1.261879321550021, -1.1657322703012933, -1.025943581524316, -0.862255409487895, -0.7054441464271923, -0.5774658054130077, -0.4885282838273457, -0.4398392218832118, -0.4230512780304122, -0.41716141398316287, -0.3989711680840034, -0.3563976580988838, -0.2886146617584754, -0.20471368063101186, -0.12123974591318469, -0.052596634484763795, 0.033699998743666915, 0.07508825836178627, 0.14329526003010154, 0.25995025674455496, 0.4137624225830126, 0.5663950210849067, 0.6834393639558736, 0.7655784384234868, 0.8286534376765047, 0.8830713693180369, 0.9255581817150443, 0.9526412875915529, 0.9557164768879566, 0.9361321996032861, 0.888818894483527, 0.8109556090336564, 0.7070624643110085, 0.5855130130470946, 0.44452673917858204, 0.2988832165489796, 0.15475703470687022, -0.004828498683600645, -0.1981406240626531, -0.42835083183802714, -0.6753699193314566, -0.9108474072753616, -1.1265010064043277, -1.315380364592804, -1.4527026365144504, -1.51743378794093, -1.5080953735741705, -1.4404052655253516, -1.3295206117378444, -1.1850714530451323, -1.0144313761861536, -0.8327929629419077, -0.6608889130937164, -0.5152003818873754, -0.40845985842470955, -0.34522089200780204, -0.3181027747704359, -0.3106061770575144, -0.30079553549581434, -0.27349572487955126, -0.22107754041210528, -0.1513027936280338, -0.08169268809122465, -0.030551303060476653, 0.005985087722671267, 0.023436905120085765, 0.07661928694276146, 0.18380691744481237, 0.3339601700885847, 0.48903039103606133, 0.6090282364047113, 0.6890971786494966, 0.747866460505058, 0.7980328514613839, 0.8359556785735764, 0.8613054414432851, 0.866787628679217, 0.8535163417340261, 0.8137073254200271, 0.7372967800466752, 0.6214981684260518, 0.47323383704635774, 0.29989298304192474, 0.1231268643446525, -0.04921830308891566, -0.23700664991769999, -0.45740799421299627, -0.7089903421636586, -0.9663127063394968, -1.2003327998700668, -1.403788450343058, -1.5666217216908092, -1.6658707560648665, -1.6815563838285392, -1.6146565526179077, -1.4831277768594, -1.3121892772576482, -1.1205469462253694, -0.9230164923448908, -0.7315666542415786, -0.5548351695486085, -0.3990937508866802, -0.27902465204915505, -0.2038427058461919, -0.17044214928965065, -0.16654086159317938, -0.1713822755141904, -0.16764509597397845, -0.13818891605382688, -0.08833992237382066, -0.037395380526550275, -0.00640266878731473, -0.012717669026532173, -0.007985418764628584, 0.038098990139981456, 0.1353718013067039, 0.27211618771500884, 0.41734467859124397, 0.5340801952077282, 0.615063812187329, 0.6723090309557802, 0.7167757251190003, 0.7475812524877115, 0.7697414609328049, 0.7787859384280945, 0.7709309488871936, 0.7336419466759606, 0.657584892532622, 0.538291194332605, 0.37997581236073374, 0.1918817939818532, -0.004139886172336987, -0.20041900812974994, -0.41303934704351386, -0.6547558093761615, -0.9183797125960738, -1.175284679476039, -1.3981975200819454, -1.5789862549265035, -1.705827057245661, -1.7607629667235045, -1.727320396991586, -1.6076624894318232, -1.421284377725513, -1.2002398931633351, -0.9710413750588247, -0.7536234161163667, -0.5587142643698046, -0.3872681155922901, -0.23456110594798205, -0.1122974666070663, -0.034740451356553556, -0.004061987276998207, -0.006986955685007639, -0.023661417866804254, -0.040701646584626855, -0.03584004814443468, -0.011056759585116394, 0.014980210614862826, 0.02084902340748609, -0.023920673396111418, -0.023929666234256672, 0.019660420036498743, 0.1083267170636716, 0.22905383183052982, 0.3599863170292868, 0.4745736484141307, 0.561236962130285, 0.6204181595492497, 0.6594752634990142, 0.6842172097244071, 0.7037770282184715, 0.714024400307354, 0.7042103934676758, 0.6628677801390922, 0.5845674039856175, 0.4658368851251656, 0.3093305965360581, 0.12095430776555684, -0.08301240744996898, -0.29541174467043163, -0.5237190753859066, -0.7735965254414969, -1.035144350715437, -1.2780377750240095, -1.47972415087839, -1.630258874670669, -1.717414083519974, -1.7270620834368955, -1.6470479490161511, -1.4815785076675343, -1.2545407960708106, -1.0012918681325695, -0.7502562832348046, -0.5233361065659506, -0.33247227515082195, -0.17429859876803208, -0.03720591499562378, 0.07176910636788954, 0.13786354424930788, 0.1580713377383566, 0.14722300918519482, 0.12386931754856642, 0.09433061125939077, 0.07588154536665691, 0.07302383381296121, 0.0716287459035309, 0.0495785094557459, -0.025721617728406084, -0.023851539136207148, 0.020086694128216398, 0.1023905541671558, 0.21081143605725655, 0.33226724678808217, 0.4497804315121435, 0.5474054659750252, 0.6125233805922302, 0.648700247067533, 0.6693046331313276, 0.6856652370238738, 0.6922098627940854, 0.6770928906187808, 0.6293081461019768, 0.5462105447133648, 0.4273271336761199, 0.2731652526708331, 0.08518420578710634, -0.12513700546773582, -0.34981039924198304, -0.5835629414309353, -0.8242544168687467, -1.0625927432909616, -1.2726869551613536, -1.440016153780109, -1.5548127176337778, -1.604600754285135, -1.5751618308008153, -1.4572376556432023, -1.2583197804416022, -1.0067140675043784, -0.7387678637366218, -0.48378011826125145, -0.2617899648076814, -0.08420461008547254, 0.056301878541396004, 0.17069325635599775, 0.2531927045578935, 0.29421660231328395, 0.2975931746416367, 0.2766838285803025, 0.24674239393428618, 0.21010811333802482, 0.17397660405521248, 0.1476447602442843, 0.1217055691967925, 0.07408566665741692, -0.016239562219197513, -0.004945567611109992, 0.045245774127722314, 0.12664228757011453, 0.22916672791505546, 0.34726220771592486, 0.472450827330014, 0.5852420383497414, 0.6624501758156397, 0.7014129530931882, 0.7203490013992959, 0.7308488087160615, 0.7285715725951706, 0.7045372690892586, 0.6465772477723755, 0.5527703266354002, 0.4270097940700107, 0.26706109353531904, 0.07319085742111356, -0.14669952629360503, -0.378218563439756, -0.6054130845094544, -0.821365939525093, -1.018066838308219, -1.1777123007044796, -1.295857092795582, -1.3666820533200603, -1.3780049918864974, -1.3170350507361248, -1.1753391130025201, -0.9627541089612686, -0.7076143326604303, -0.4429087652670136, -0.201329582785157, -0.001585177293534679, 0.15082991086066364, 0.2660330720179948, 0.35089389569690194, 0.39969754310861344, 0.41037541436448177, 0.39426846746999467, 0.36358973320244475, 0.3266517713358748, 0.2837931040663235, 0.23656628277457814, 0.19461475528914562, 0.15014124861109965, 0.08536172036484452, 0.0010114137179957437, 0.02941054268665454, 0.09349780510209015, 0.1822799231475269, 0.2876991005254485, 0.4115212763573947, 0.5478020300104459, 0.6747391525144492, 0.765636651236314, 0.8128939847909573, 0.8345448786555972, 0.8430603670886481, 0.832110609077286, 0.7921513479662338, 0.7143649117323257, 0.6010559398993085, 0.458939774189711, 0.2843442209088753, 0.07734395030571993, -0.15221437663336995, -0.38300113715574235, -0.5938665333577499, -0.775186165755206, -0.9213284073977211, -1.0237219572836969, -1.0851421819096965, -1.104420390102205, -1.074952914144542, -0.988627983713394, -0.8364888146087903, -0.627413175019425, -0.38778179852038547, -0.14420558073969286, 0.06909434838744054, 0.2333232698258123, 0.3479239241385424, 0.4250783970046437, 0.4705288386528189, 0.4811127933052884, 0.4599279024310807, 0.4228270814572225, 0.38155253366564745, 0.33749513939034026, 0.28850881348069907, 0.23571831961599296, 0.18887564383798805, 0.140089293822383, 0.07654922802346431, 0.028308980699517594, 0.07982739370285576, 0.16302759830266322, 0.2673958187083631, 0.38612190555466847, 0.5235704540197914, 0.6725100986608693, 0.8099521665168571, 0.9122303049137304, 0.9714952787789358, 1.004824499295779, 1.018041624574052, 0.9987436107205914, 0.9358343741777247, 0.8285228435894527, 0.6846785227991733, 0.5166206613359983, 0.32103817135233625, 0.09857330327240395, -0.1359812334782295, -0.3575042025879996, -0.5443696669463224, -0.6868526155173446, -0.7842150724211037, -0.8339227487991022, -0.8398624849921175, -0.8055254934351302, -0.7367433948665773, -0.63070979331493, -0.4774869640345505, -0.28581269736779513, -0.07712916011316831, 0.1283439727062784, 0.29921236761835157, 0.4188031377455466, 0.4885307021088217, 0.5188710114177878, 0.5181155897621764, 0.4886036849064472, 0.4345834852348993, 0.3723849313233728, 0.3141392982901736, 0.2591787785224003, 0.20379811379848542, 0.1501491913734552, 0.1107347362480321, 0.07911917657528968, 0.043433027747960574, 0.0601402241966695, 0.14018386385307563, 0.2483846741928609, 0.37629020583980477, 0.5189200772944628, 0.6777910415861795, 0.8391377474269629, 0.984042040697186, 1.0969761409232985, 1.172099071362534, 1.219965615240487, 1.2363260075258147, 1.2030971457408621, 1.1096330039432767, 0.9647113968635364, 0.7840197285467748, 0.5845976944741007, 0.366568308900666, 0.1332500494260123, -0.09838227115647027, -0.3028115739024069, -0.46062173343556184, -0.5649676817544808, -0.6192618864510109, -0.6250480971086916, -0.5839132168164577, -0.5021566970062814, -0.39764845820012334, -0.27565024009351147, -0.1273962767039235, 0.038984695548062595, 0.20726914015453707, 0.36430409761833576, 0.4842497673840951, 0.5548409994420122, 0.5770261636158975, 0.5584001457801099, 0.5100097208305739, 0.438541330420349, 0.3488020004929823, 0.25677114040687266, 0.17432000250734825, 0.10191977396076914, 0.03811266119696804, -0.010558284680941223, -0.0305138126914014, -0.026740563436968578, -0.01228653728180135, 0.08668840948553495, 0.1945541434235551, 0.33131479435651695, 0.49066242630348256, 0.6669253998083869, 0.853587091691582, 1.030738090542012, 1.1804203144798837, 1.2990598312231267, 1.3872852761692445, 1.4459476444529515, 1.4597960424139964, 1.4085228169954236, 1.2821843498021537, 1.0977332043275174, 0.8787922974058104, 0.648119355685435, 0.4111984719631688, 0.1730260514947934, -0.04889122309139184, -0.2296738445385273, -0.35585570819934204, -0.4238525629514863, -0.43881034732439905, -0.40666484465630076, -0.3282312898994144, -0.21230892754465747, -0.07917795522756765, 0.05786050218463522, 0.2001813246063916, 0.3385784509363245, 0.46362613806068836, 0.5695681824356945, 0.6364834901133382, 0.6557903070601097, 0.6290843746620072, 0.5631667937383041, 0.4690801819624446, 0.35453617271822035, 0.2294391182933254, 0.10877696477341114, -0.00017808352086527346, -0.09412739190547555, -0.16914519071034922, -0.21232987799028, -0.20876438876260275, -0.16057860145515626, -0.08345207098208728, 0.11068739514107295, 0.24516125798460348, 0.40983100084624113, 0.6013487269507924, 0.8106933767116864, 1.0220840465720682, 1.2139406127730559, 1.3686200113044669, 1.4895410915278742, 1.5825148304936625, 1.6413140497427248, 1.6458013625387002, 1.576059367149562, 1.4229204093483219, 1.2075069825870979, 0.9587150939663233, 0.7034262994700478, 0.4525522931144074, 0.21452595293629187, 0.008518683792221295, -0.14572700162492544, -0.23909606300687336, -0.267851007661348, -0.24155109821411955, -0.17319521363941684, -0.06816984971773793, 0.06600082965459524, 0.21367367150888994, 0.35939656245218327, 0.49595723608668424, 0.6115708702428493, 0.6995324509434478, 0.7555788130116878, 0.7656911276623998, 0.7295287450690109, 0.651972055538641, 0.5408785008839522, 0.40626020899905935, 0.2549344812651705, 0.09857552568203043, -0.048321541045871615, -0.1815304111761185, -0.2944177563476078, -0.3786372945912682, -0.41552538839522246, -0.387272701447139, -0.2951336637381248, -0.15650365194491064, 0.13275309424779902, 0.29048393116438387, 0.48113424806977634, 0.7004246804792283, 0.9334994618549791, 1.158909584651125, 1.3605472203212379, 1.5233877119977175, 1.6485424471344998, 1.7389301798177659, 1.7874331314539749, 1.7757149603002478, 1.6886010504129538, 1.518046800212621, 1.2849450524375732, 1.0184092361129096, 0.7493136464776203, 0.49584907044202675, 0.2689932798087573, 0.0831645233412847, -0.04658066481448362, -0.10739271867023402, -0.09566523059418448, -0.028563358264520722, 0.07211343162422829, 0.19276420698971317, 0.3294063934261113, 0.4748520365675091, 0.6161844809767539, 0.7439824398499404, 0.8439680827732434, 0.9058868091094369, 0.9206935808133941, 0.8785083577456125, 0.7870227897808098, 0.6585809435608375, 0.5043600702745075, 0.3327817757687217, 0.14965173322857286, -0.03368968234557732, -0.20399079597620895, -0.3565673597685923, -0.4808252169733748, -0.564890313806834, -0.5880042910644832, -0.532865843672339, -0.4022278600671061, -0.21460628872105292, 0.15171570131701145, 0.3294766279588963, 0.5433802290429551, 0.7843044937191996, 1.0289784637804713, 1.2563355880827485, 1.4608519579460908, 1.632673324754094, 1.763400593893932, 1.8482857640946955, 1.8814625900824438, 1.8520331936304035, 1.7523421974674749, 1.5766723920683368, 1.341008452669209, 1.071202206852692, 0.8018781729132651, 0.5596005625612447, 0.3560165228743097, 0.19657076722808703, 0.09196288005699438, 0.05701949427746135, 0.09623015151282824, 0.18915954798868803, 0.3069306406872845, 0.43045924108851086, 0.5577325226695207, 0.6890047635072076, 0.8162185905071667, 0.9313860747521422, 1.0218766579275935, 1.0704329973176812, 1.0595054215284136, 0.9790768874000363, 0.8421595911651162, 0.6670262974133523, 0.4700866333984316, 0.2624705681439188, 0.05011623404914545, -0.15670519300974392, -0.3467499052287073, -0.5105276519613462, -0.6341697419681395, -0.7076950254204044, -0.7122915882092311, -0.6340004243290305, -0.4755702331276026, -0.2550795255817335, 0.16743167131338557, 0.3638967395337129, 0.5962154457571484, 0.8475062525502227, 1.0917171596839994, 1.3131362901723453, 1.5147461066195997, 1.6898335711492287, 1.8224205385431325, 1.8985551819352842, 1.9136359544151165, 1.8681710270092242, 1.7627728506120803, 1.593233724262754, 1.3708682271201846, 1.1178233631140262, 0.8702299714560348, 0.658986851876473, 0.49402146331469693, 0.37048721629778963, 0.2918634840035892, 0.2713416250532549, 0.31537986187017114, 0.4059862648865496, 0.5164723967172787, 0.6288529875436024, 0.7392106424731831, 0.8490934736926877, 0.9556599942033016, 1.0561632689253317, 1.1421578879811993, 1.189393325401079, 1.1697640563339184, 1.0704068149490464, 0.905176931790449, 0.695131364125613, 0.46127938047430145, 0.22002190931954813, -0.021471439986458733, -0.2529315546899461, -0.46242390401879885, -0.6339560772427788, -0.7511605564299271, -0.8085434238308501, -0.7904379610720424, -0.6903263985169439, -0.5133335588080877, -0.2754314730832395, 0.17858243598148782, 0.3858165672703741, 0.6252141685158843, 0.8741642691551188, 1.10809159086088, 1.319720655959584, 1.5151063750327747, 1.684523921716817, 1.807886186433704, 1.869439180317817, 1.8671258412609333, 1.810108198719666, 1.704812167444125, 1.5506310682489568, 1.357283094728669, 1.144008222084117, 0.9454118105105371, 0.7911002472875507, 0.6832142275892423, 0.6034884443935066, 0.5474873757575638, 0.5275721752611897, 0.5535898254945593, 0.6177643470663685, 0.7039795308716877, 0.7942221106222881, 0.882907339490559, 0.9676974970548037, 1.0485882940704672, 1.1316596187057386, 1.2164446172241938, 1.2703360433124187, 1.254442500415316, 1.1522448272688892, 0.9742782760199445, 0.7427599791131866, 0.481863292640154, 0.20977521567779353, -0.0611165929942359, -0.31572757289899284, -0.5409298613410418, -0.7174950912368838, -0.8274114182793486, -0.8676949828688006, -0.8265596764550127, -0.7062453170403021, -0.5174650086483688, -0.2746904215902581, 0.18111603647984262, 0.387994050217292, 0.6203080225717813, 0.8546654514655714, 1.071313773382841, 1.2672056950474564, 1.4458928276736853, 1.5955262153968015, 1.6994134593960128, 1.744246000899542, 1.729402671643084, 1.6663803873164087, 1.5646188281289604, 1.4299020682106112, 1.276637033110952, 1.1243007726352097, 1.0009641066926016, 0.9271313401962449, 0.8921645780646232, 0.8611171513615132, 0.821566134683544, 0.791872519442839, 0.7915776394987578, 0.8246476492188947, 0.8815247405356761, 0.9456879447983595, 1.0085137607622603, 1.066336752040762, 1.1201915399549385, 1.1832432735192044, 1.2663085279218669, 1.3296530074514568, 1.3221378572591875, 1.2240760558730643, 1.0426284421775793, 0.7987563263689442, 0.5178658302174028, 0.2201448030180661, -0.07588876106430088, -0.34762703988759946, -0.5814328250931832, -0.7578720227305958, -0.8596821426111686, -0.8854374389121824, -0.8268382731281818, -0.6936431115187606, -0.5019746243961503, -0.2640058963475149, 0.1729657378534728, 0.36639438792424056, 0.5776502038775692, 0.7881398649529447, 0.9808675966157645, 1.1514702779605632, 1.2999192737473313, 1.4173407733924384, 1.4943346300653721, 1.5217927110612546, 1.498171677342343, 1.4333050848350877, 1.337959956422552, 1.2260010114676847, 1.1181333094489032, 1.0365397137912056, 0.999565645179918, 1.0139485796474474, 1.0563176558522684, 1.0771814365201597, 1.0557042321396983, 1.0210099010281157, 1.0053238898348453, 1.0200842547526237, 1.0548081755084024, 1.0953131420906086, 1.1326727632546072, 1.1667720085276954, 1.1984455387793123, 1.2428583577954597, 1.3212458423855848, 1.3898071992954049, 1.3873970700786136, 1.2910733994594545, 1.1077039156010964, 0.8559312531734501, 0.5592486243527053, 0.24092385659923574, -0.07422388639546547, -0.35525758826996706, -0.5899281409871869, -0.7635035004456694, -0.8574049095724725, -0.8689651945593044, -0.7971587821629861, -0.6599256957667244, -0.4726519341914025, -0.24578080601303864, 0.15666489260643293, 0.3252021226695726, 0.5039029760316337, 0.6798513148141848, 0.8402408840789529, 0.978747982147306, 1.0903196109710007, 1.1687248791693494, 1.2133556163366377, 1.2209865251863787, 1.1904505783940433, 1.1281874874422249, 1.0449397193361527, 0.9583440276648134, 0.8943015792903419, 0.8792081424728938, 0.9210008390313049, 1.008710580514286, 1.1109633973707749, 1.1789380406349879, 1.1855388039131356, 1.167279621746697, 1.1606867332184296, 1.1778898328219503, 1.2082224674372863, 1.2386405795478406, 1.2631336652452405, 1.284452054014401, 1.3047238911591132, 1.3408446779117793, 1.413408236194487, 1.4768178267392664, 1.4686795419159933, 1.3681814568710444, 1.1822304091216524, 0.9269938653199558, 0.621006968587551, 0.28897746615654885, -0.037763160638784966, -0.3232857416263372, -0.5554574123859632, -0.7260263827153917, -0.8164474073956167, -0.8213575128122508, -0.7444489212341631, -0.6085153078843981, -0.4304855242411306, -0.2211951630815247, 0.1330784730435229, 0.26973812759481897, 0.410268389945964, 0.5453286331532994, 0.6668914451622946, 0.7667170504795175, 0.8350958779801277, 0.8688544135303674, 0.8751503608067736, 0.8593226760627891, 0.8228791781818453, 0.7688563947570477, 0.7055402299229451, 0.650136412426943, 0.6286121232557177, 0.6665072867568225, 0.7619622626195061, 0.8909583745056187, 1.0241999634184584, 1.1276413219420838, 1.1772764663697768, 1.2010912689243225, 1.2319462969661514, 1.275941251152753, 1.3259366198315197, 1.3707673850788493, 1.404979176221387, 1.4306316701019925, 1.4557078592988677, 1.4971772311344596, 1.5646765966171683, 1.6142274411297945, 1.5903693369667278, 1.476970241745646, 1.2826259734470455, 1.0241209345815712, 0.7155020373223742, 0.37960014043103574, 0.052256946029102566, -0.2297697632610315, -0.45662825955979663, -0.6270225819908898, -0.7227146690333145, -0.7339371333471341, -0.6666484546650709, -0.5419878133248535, -0.38058703988319537, -0.1948448147869205, 0.10137106473957637, 0.202941808119985, 0.3050833614617257, 0.3998550820307678, 0.4798854003938076, 0.5349187849402072, 0.5552367107657843, 0.5410090514617409, 0.5054584130560965, 0.46362564909684983, 0.4203564218198092, 0.3756507197910725, 0.33630761422675953, 0.31828548200401086, 0.3403696425239063, 0.4180145341701932, 0.5419666305403541, 0.6854655403173956, 0.829277817389202, 0.9578240735298118, 1.056634064549885, 1.1387507350993065, 1.2249970076968062, 1.3141314368371355, 1.4021446132516053, 1.4817732239304355, 1.5462951457072416, 1.5954543668385783, 1.6390072995756635, 1.69216594717542, 1.754992309664196, 1.7881678851217464, 1.7472264334984609, 1.6187971967734323, 1.4127071473645811, 1.1482903481098135, 0.8407453896785732, 0.5111430925968891, 0.19310413617516264, -0.07893361118158214, -0.2980768446526025, -0.4691549731885652, -0.5765699712015339, -0.6058788408050082, -0.5611069505030971, -0.45923526899779366, -0.3243998630189301, -0.16832992675475356, 0.06528861793152492, 0.1321620633330085, 0.198864340160087, 0.25697330670097823, 0.29555910023706006, 0.3048770820977645, 0.27868202629191885, 0.22152764315598958, 0.14839075899348167, 0.0813027266654698, 0.028546765475160395, -0.008652678698300671, -0.024889435191334536, -0.006883819752326106, 0.05348464533559039, 0.1587550336744191, 0.29505060929109794, 0.4427726434083275, 0.5941726268530774, 0.7433174532745247, 0.8856569399233222, 1.0266185382733473, 1.1733411198830097, 1.3161979941399344, 1.4514051728133355, 1.5732190187990005, 1.6748630729001233, 1.7536958652625818, 1.8175169543399203, 1.8816019041376468, 1.9421176328310266, 1.966002572015339, 1.9165706743861792, 1.77831622742713, 1.5629862189724237, 1.2943547082877667, 0.9916017789357122, 0.6746343030068955, 0.3736782090235047, 0.1172627720299357, -0.09309865553381881, -0.2654363523010491, -0.38646674265550024, -0.44049607479387726, -0.42706677552373823, -0.3593950015658286, -0.26005617388222424, -0.13848702298322596, 0.03197082471588381, 0.0677741807681973, 0.10440886300168593, 0.13178650989593044, 0.13335280461942206, 0.10155728253200673, 0.03523520749881175, -0.057702922783241004, -0.1621567724739608, -0.25205570796873744, -0.3153122883872628, -0.34756099456042167, -0.3430860855547015, -0.29527070990358384, -0.20793853621888714, -0.08779055602399377, 0.05294676287122044, 0.204113442040026, 0.3640931038472579, 0.5304950292093219, 0.7064804911366511, 0.8989970048061194, 1.1040861277853458, 1.3017271209802972, 1.4847430340160688, 1.6469231333324057, 1.7826788667256523, 1.887293235527346, 1.967041648668365, 2.037694109537794, 2.0964510703596733, 2.118548313618303, 2.070546019739245, 1.9343626501368485, 1.7220959826209663, 1.4578502922407668, 1.1650304471788646, 0.865089563914735, 0.585572380259084, 0.3468662407767608, 0.1457481030151439, -0.027416142103110514, -0.1628607019600312, -0.24574730260698138, -0.2709943544530861, -0.24728097310299524, -0.19043600471410205, -0.10628878321110728, 0.004464116372067288, 0.01477511648304728, 0.027975445598795994, 0.032397834132197316, 0.004764625763097604, -0.06032068271599778, -0.15940010632166954, -0.2829392376913925, -0.41405718053567886, -0.5232065122604864, -0.5952478159659937, -0.6236457705807389, -0.6044780374797987, -0.5389651509146822, -0.4376820299046751, -0.310056181488616, -0.1641047467624552, -0.005394648677599143, 0.16339281308051637, 0.3421887623456822, 0.5416912821106262, 0.7740262994171043, 1.0288662482243183, 1.2770044755223722, 1.5018838497952272, 1.6966389334106557, 1.858622426129689, 1.9831204907502071, 2.074481733598019, 2.147492573325409, 2.204610752993006, 2.2283251528593278, 2.1879633200998314, 2.0646195500935276, 1.8695618810458507, 1.6235752789158724, 1.3497684281721367, 1.0719279480722594, 0.8133660952354981, 0.5882341193209172, 0.3926908690373209, 0.21808169411586784, 0.06892585687141681, -0.043186181958821415, -0.11072284128222781, -0.134367117047214, -0.12111682183460362, -0.07405043318692352, -0.017518842737226984, -0.0275369917063503, -0.03183634470252937, -0.042740777760290026, -0.08792594520975522, -0.1739672901371283, -0.2984345078183025, -0.4511768940534909, -0.608150595213415, -0.7370769755861221, -0.8197338054045378, -0.8473721634244598, -0.8194623014085656, -0.745074330754205, -0.6352336203355979, -0.4990936005271469, -0.34246378311511383, -0.1725758693506014, 0.0038429824097336454, 0.1881437030079003, 0.39787249597649504, 0.6523547796800152, 0.9415480860505822, 1.2320118465129895, 1.4926225498463663, 1.7118576416347886, 1.8917621182923752, 2.0314120031935103, 2.131525814943559, 2.2036645584423415, 2.256927235910617, 2.279728550641538, 2.2487009171621546, 2.1455538331142074, 1.9786446562057693, 1.7645770539317394, 1.5211025845228017, 1.2708498941367956, 1.0317882967981347, 0.81502101471053, 0.6196071428640066, 0.4397204156510659, 0.2767869127006417, 0.1400895107083132, 0.03712424438110786, -0.026656559522865827, -0.052271317654839465, -0.04056729580171091, -0.03382673433122877, -0.0586665557728639, -0.07302595558256023, -0.09137481260284627, -0.14314565668604998, -0.23981380304885155, -0.3851331899495692, -0.5671443620227862, -0.7515315417583773, -0.9019643350472255, -0.9984733551242997, -1.0290503145279954, -0.997039679044038, -0.9155691069830615, -0.7952951581829811, -0.6487370466306531, -0.4808769100034731, -0.30144540800242325, -0.12069815113550639, 0.06382097420682392, 0.2737332420874411, 0.5345838259159111, 0.8408089054086502, 1.159394803660957, 1.448846197356315, 1.6880492440654078, 1.8819254760556323, 2.0339883116944173, 2.1403312232952767, 2.2065138901692234, 2.2476178900405053, 2.2633607477759083, 2.2388950944620207, 2.1567882924757997, 2.019312774707032, 1.8406189732689067, 1.634037984769709, 1.4177950190343211, 1.2023557225202128, 0.9956432810617868, 0.8001179871780253, 0.6131304792234786, 0.43706289992576053, 0.2838325323890222, 0.15887703999263547, 0.06817653912011142, 0.012795674311130573, -0.006753917991849055, -0.041039712362719055, -0.07237178916965846, -0.0892140912504788, -0.10926153010307417, -0.16026306873206142, -0.25955699255553594, -0.41819175944510234, -0.6236546898011045, -0.8310105031971482, -1.001230380750802, -1.1141163548257982, -1.1528895435579194, -1.1233047471692215, -1.0395700067298557, -0.9102395176250482, -0.7556005803568665, -0.5827639013895226, -0.4002368039381114, -0.21791876256563122, -0.03304368661080486, 0.1759523291297765, 0.4344402178683602, 0.7422182505461575, 1.0704758272060644, 1.3759025516216095, 1.6288969073906614, 1.8327128193686064, 1.9899703904532058, 2.0952846380840766, 2.150786623917122, 2.173889113688993, 2.17683567385132, 2.154635899953814, 2.0903127316079346, 1.9784323841945446, 1.8308734362722454, 1.6594936392227129, 1.4783135440604098, 1.2919649986440067, 1.1035630811098471, 0.9167249841111423, 0.7315254632697462, 0.5518476629931016, 0.39315489085083055, 0.258747628048281, 0.15382618423753538, 0.07845376196318218, 0.03032100953453579, -0.0337762441172042, -0.060050456767751545, -0.07348075228264637, -0.09245685330282827, -0.14196546841045324, -0.24129918988297896, -0.40467384014062896, -0.6216862124729857, -0.8425948968254681, -1.027625138427754, -1.155379706907037, -1.2070222439536562, -1.189679787595759, -1.1123137316551814, -0.9803628761446402, -0.8232093101410579, -0.6532185768219245, -0.47462235406613645, -0.29142858787449927, -0.10259827511611772, 0.10858974940801645, 0.36071499131214674, 0.6580378569602022, 0.9792515650567037, 1.2843637337831546, 1.5398091808574303, 1.7424074922771726, 1.8924129023381577, 1.9861888086815338, 2.0285074697294094, 2.0349958915838653, 2.024812049824504, 2.002052071969609, 1.9506368231359976, 1.8592962523767036, 1.7389151741405418, 1.6010450169922328, 1.4573534645719706, 1.3064707309804287, 1.1459253650655283, 0.9771391131594394, 0.8056943727926708, 0.6361217830272867, 0.48380951185736826, 0.34970574994200776, 0.23756270658608944, 0.14582390632373068, 0.06917685139598251, -0.01147677086065401, -0.020237496903397715, -0.026304127981107376, -0.0451317959161097, -0.09569884193594719, -0.19468133203259685, -0.3569798341651564, -0.5749305627241839, -0.7999199208830048, -0.9946297040866334, -1.1350998173158195, -1.2036272765541345, -1.2053783643498235, -1.140141408459053, -1.0130295304629493, -0.8604125211699267, -0.7001509219109232, -0.5321857157520287, -0.3496434213613979, -0.15180286844238638, 0.06447167975348088, 0.3069217602352998, 0.5839211775113495, 0.8842879397906607, 1.174380665089464, 1.4184474952437611, 1.6044702290118635, 1.7343646227784295, 1.80925540466866, 1.8362546472004395, 1.8295666083800262, 1.8090623630627443, 1.782925082812088, 1.7385136423073715, 1.665659791159182, 1.5721058295405028, 1.4691986195066211, 1.3652517734526453, 1.2549098847937874, 1.1313139991173147, 0.9932294335343831, 0.8477623709416442, 0.6989254744419204, 0.5580935417704935, 0.4271622391256265, 0.3093195187879999, 0.20338277679560005, 0.10218932822308605, 0.020811790601629555, 0.03753009303367894, 0.04278802834863486, 0.02481658735892021, -0.02819454360113651, -0.1276333887707587, -0.28494381588733914, -0.4952837904122178, -0.7172569556750678, -0.9177476585203437, -1.0726189453388812, -1.1645785846708958, -1.1915973700856632, -1.1431638359249439, -1.0273193041059772, -0.8837201680054306, -0.734560436678524, -0.5760007815273842, -0.39329429157252715, -0.18603010383313467, 0.03457991031664227, 0.26507904439852453, 0.5154685833147602, 0.783207231000417, 1.0434719531561023, 1.2612015671690502, 1.4182333303354602, 1.5208583712195833, 1.5752752264416447, 1.5881256955130927, 1.5705125954757824, 1.5396075902842503, 1.5049786151291769, 1.4630608513210035, 1.4071071006525422, 1.3404173429901811, 1.2719917190768182, 1.2074126483368337, 1.1405204343933963, 1.0622912943754639, 0.9678825678391231, 0.8590742590524234, 0.7361903272028667, 0.606793964528494, 0.47842676838353093, 0.356293555750393, 0.23980548914354574, 0.12219234641046896, 0.05376008621723068, 0.0974858799290242, 0.11741928825886624, 0.10346070662855845, 0.050511440608947265, -0.04610138622776918, -0.19369447580251006, -0.39194989315079176, -0.6083859794822385, -0.8134457088802697, -0.9826346074453323, -1.0988740185457142, -1.1534617130954887, -1.1291631544152707, -1.0319774236083128, -0.8987031881973392, -0.7565107608494771, -0.6018506332811265, -0.41813497192977056, -0.2065715293542107, 0.013349638352556441, 0.2307676748497435, 0.4540178483307702, 0.6832039865725523, 0.9010496632682217, 1.0772069493111056, 1.1944946238598382, 1.2655234043365058, 1.3017339073881842, 1.3059507716503238, 1.2826384509506843, 1.2404020184207711, 1.1924380314555276, 1.1472357867615437, 1.1040152171177517, 1.0620824786964298, 1.0274184792358416, 1.0023218276137402, 0.9815287202631431, 0.9525495778917448, 0.9065987709309415, 0.838277745451919, 0.7441496314612118, 0.6283428699909754, 0.5045190447841109, 0.3798776055560894, 0.25618274913430783, 0.13039789773390661, 0.08051452891448664, 0.14728952348956206, 0.18239855468144414, 0.17507899195763016, 0.1251548565397949, 0.0358399937706483, -0.09805718457264323, -0.28288017850296476, -0.49314997518920084, -0.6992736256001479, -0.8747022014407928, -1.0042427384967378, -1.0782083163357579, -1.0798447901130022, -1.008441718574366, -0.8913312900899302, -0.7555281901719587, -0.6035190392794646, -0.42232798762306967, -0.21393702722404387, -0.0009381476756278562, 0.20238288527540854, 0.4009413100787779, 0.5922137829931452, 0.7627452919961452, 0.8891964815599922, 0.9595347157785229, 0.9951640595105385, 1.0130278430376967, 1.011727433754297, 0.9863187012724669, 0.9369058234738715, 0.8790937723160528, 0.8307377902475559, 0.7966046382539318, 0.7747385280487253, 0.7686084674962254, 0.7798308689849364, 0.802195486861125, 0.8209326391024617, 0.820830069023133, 0.7910473795815551, 0.725060597544478, 0.6261383507510309, 0.5112561561647477, 0.388257907688608, 0.2615533224329588, 0.13286006614430898, 0.09968480891310177, 0.18240857081309517, 0.22783053307698165, 0.2257500434624118, 0.1788385321769669, 0.09647389885230682, -0.023364608437922124, -0.19152013409666216, -0.3909405014226646, -0.5905829164277911, -0.7613709235915159, -0.8886220276115352, -0.9680932000382048, -0.9912080123148599, -0.9492021419041986, -0.8554490662074938, -0.7315125772814944, -0.5864173041591516, -0.414845370993351, -0.2177486258389366, -0.01715730111426026, 0.17246229888485728, 0.34974850606820895, 0.5080880238203982, 0.6337348817004609, 0.7094273952735803, 0.7325992406305057, 0.7319128433470485, 0.7283057294767883, 0.7175216693843774, 0.6889411876342644, 0.6400959564619829, 0.5854713574190601, 0.5424467230679559, 0.5189403631478975, 0.5120923818472777, 0.5257572357111026, 0.5631458749140693, 0.6202918848659498, 0.6789524528422249, 0.7166700841079883, 0.7186571140139967, 0.6789883004418975, 0.599841895469696, 0.4986836316888839, 0.3841299391227331, 0.2624470775361669, 0.13444110623073383, 0.10909582584874827, 0.19935735244675426, 0.24945577889702558, 0.24903404404075336, 0.20227804129295074, 0.12265984671746537, 0.01420512446995673, -0.13522144184974497, -0.3159425483640898, -0.4973998125333571, -0.6528035113000996, -0.7682574207447215, -0.8439033380890958, -0.8788801695292329, -0.8601305607890721, -0.7893842122392823, -0.6822078806522219, -0.5520962908830833, -0.3992159421350376, -0.22279269446435176, -0.04053252865685856, 0.1328246212040807, 0.288015566585365, 0.4155141101140108, 0.5019970926743289, 0.535042847987852, 0.5201970463142847, 0.48891200412887753, 0.46328781285626014, 0.43888000188526033, 0.40485830724262917, 0.36228668068540537, 0.32072639887990617, 0.29269533571687295, 0.2824312526353208, 0.28723991598460497, 0.3133261813868225, 0.3668038537084719, 0.44561540599797894, 0.5313779195790072, 0.5965518222754355, 0.6235483213167565, 0.6072538450791698, 0.5499715094384147, 0.466313799255263, 0.36591473511490336, 0.25655000674670203, 0.13421652604974962, 0.10488615684399474, 0.1935410878649296, 0.2452331249121634, 0.24774344498201148, 0.202987230963773, 0.12759185539117288, 0.03125406376977949, -0.0977706776427982, -0.2543037211384459, -0.4108613927817349, -0.5449812889755504, -0.6459940634887973, -0.7128224800048334, -0.7485997943321357, -0.7435196228664144, -0.6927469414038163, -0.6057691724283503, -0.49793030764907975, -0.3709658035612827, -0.22311820935299084, -0.06803166076373832, 0.08142514972041784, 0.2112398815829638, 0.3096539356966786, 0.3660544308008899, 0.37375974573632964, 0.3392758763201728, 0.2880673043871023, 0.24054224817158718, 0.19696276988521763, 0.15581771138193087, 0.1219417374135179, 0.09716522122774647, 0.0876141617135912, 0.09132606069320508, 0.10478170172926404, 0.13879695881282894, 0.20030960897019823, 0.2875014637121715, 0.38605671080057535, 0.46811021358504346, 0.5134694537663901, 0.5165448365126828, 0.4797378266865502, 0.41560267415353974, 0.333690231881742, 0.23949522676487578, 0.12720503969063202, 0.09092768434760687, 0.17280800198328916, 0.22407196327301587, 0.22924531267427842, 0.1899842682144223, 0.1260053593779553, 0.04877036312146823, -0.05608489706126985, -0.1863361945723788, -0.3185330959140506, -0.43160255425015737, -0.5186484544399163, -0.5746045949200372, -0.6047874186542384, -0.6063893861169762, -0.5733124916911015, -0.5083246926860332, -0.4258448092677657, -0.32834092498764506, -0.21373309357596104, -0.09065476402341902, 0.02937549175625686, 0.1303002476803752, 0.20136880828415674, 0.2379521883909755, 0.2381420175976718, 0.2024002766665321, 0.14446880750671778, 0.08071167121734901, 0.01935063819159208, -0.029453037180322612, -0.057160996918070854, -0.0700875032641203, -0.06670460012914943, -0.05244277036761878, -0.030550760941904835, 0.009898093255204268, 0.07443491354914263, 0.16004811648910375, 0.2593910991166609, 0.3469784394801635, 0.402826443808473, 0.42088306541680093, 0.40212579165334245, 0.35571420309256924, 0.2917286567491367, 0.2116138572565979, 0.1123368192978921, 0.07176346364454507, 0.141944750152663, 0.19118304980647371, 0.20152026783656823, 0.1715783828366507, 0.12051876434497764, 0.062059637271896895, -0.01720000539441763, -0.11865803461191754, -0.2249182807656654, -0.3170017507742071, -0.3877902839484789, -0.4313180884416163, -0.45320281910106414, -0.45579199821169913, -0.43411516032561276, -0.3908225099161868, -0.3367025284339979, -0.2715391717197991, -0.19018136190302842, -0.0987402368033618, -0.008587539298624413, 0.06480136821298856, 0.11324847902236967, 0.13648332776218153, 0.13623248374028155, 0.10698275458137005, 0.05182464232673978, -0.017700683630382717, -0.08794269252573257, -0.14147401192370263, -0.1680738192901522, -0.1768713557537912, -0.16769695148981714, -0.14536470882481123, -0.11517837045616837, -0.0701640731107268, -0.007213014672436979, 0.0709460085943138, 0.16028380085174534, 0.24341367085669813, 0.30299779723167036, 0.3334125403687824, 0.32930450110245807, 0.29574774112188934, 0.2443817378138808, 0.17637571863159662, 0.09221546176331061, 0.04958815637781112, 0.10236881941794056, 0.14556954069479136, 0.16120233464376246, 0.14189290770780488, 0.10056575413920206, 0.05501318078904362, -0.0017222709656934683, -0.07297956038994083, -0.14974020260843912, -0.2172411992687594, -0.26746183426785686, -0.2960891158535133, -0.30675648008700096, -0.30490868817625094, -0.29015846537985673, -0.26717469906481545, -0.24083775503604182, -0.20530159201371262, -0.15323071760604523, -0.08986696182225115, -0.026382384175520582, 0.024973356193701095, 0.05656678996610149, 0.06948005873665891, 0.06733079439490824, 0.0411940200968382, -0.008785114438591668, -0.07314434396064681, -0.1390115140141042, -0.1903831869508609, -0.2157016737864477, -0.22219078145633478, -0.21101289572804635, -0.18547505233616024, -0.14940050264474786, -0.1023190514727775, -0.04551529395413135, 0.017849923591862178, 0.08817235242987287, 0.15852189399215744, 0.21511378638702092, 0.25191076627553516, 0.2572995882562503, 0.23330667323867765, 0.1926107982087456, 0.13823066558604274, 0.07176001055877392, 0.030285864337867684, 0.06422432562025349, 0.09504100703990068, 0.10936059747148064, 0.09690049710375356, 0.06457836403575433, 0.029890740877758875, -0.00799095039950016, -0.05179606316323251, -0.09985792937394969, -0.14177567858742612, -0.16967130490757515, -0.18147219758155042, -0.181585235253248, -0.17598382536715132, -0.16738979368933438, -0.1601156452498267, -0.15387725335247293, -0.13909220436615494, -0.10820842769720264, -0.06753568671040747, -0.027235725197626875, 0.005667010048501968, 0.024451374129262623, 0.02898760684243141, 0.024264241523706712, 0.002422445168675967, -0.038028366080548964, -0.09000921902038543, -0.1420746805426034, -0.18221680855611033, -0.20180671450623336, -0.2055240999705643, -0.1952759269120635, -0.17184666172970248, -0.13745275224490164, -0.09502648626915147, -0.048501635570823355, -0.0045844633409553165, 0.04019056760431577, 0.09001610933366919, 0.13595364576307506, 0.16939769159659696, 0.17735200345571558, 0.16222510405115648, 0.13459957770934117, 0.09697145361569205, 0.05057272695266765, 0.014945429092312144, 0.03135480572008818, 0.04685328378230124, 0.054716554589370664, 0.04722013965114326, 0.027832522460065064, 0.007858047340447639, -0.01208012320080421, -0.03258755242838501, -0.05496940948350193, -0.07392539976260462, -0.08463135794684233, -0.08575540793424134, -0.0822952841097145, -0.0777099083132041, -0.07442167953676385, -0.07392353185754422, -0.07442266730851144, -0.06946670073815754, -0.05523372590424404, -0.03577634491527682, -0.016820138768919555, -0.0012976278669376052, 0.006697528904373199, 0.007352661381543478, 0.0042598883921659385, -0.008164449890100666, -0.03192108671031779, -0.06244057533772213, -0.09204226385494586, -0.11434303589567715, -0.12458798078407168, -0.12553919628549562, -0.1191382488190383, -0.1047061463917257, -0.08309165784223838, -0.05734695919725902, -0.029777836713500445, -0.007061827842902266, 0.013704466882009275, 0.03850591632316134, 0.06410418223325202, 0.08388077725389163, 0.08988280983422035, 0.08321715590108508, 0.06959072861166651, 0.05026851520197798, 0.026254234659231494, 5.524575406262912, 0.12015959380339386, 0.11987290689737741, 0.03375622886248061, 0.1364162262915162, 0.1767038743465052, 0.07625211104244703, 0.06626676581195279, 0.1389309876744938, 0.08696684653800922, 0.022209681895174364, 0.10619521459089054, 0.07522801765569398, 0.034866375713591115, 0.10672024351786144, 0.05439896790154933, 0.04213221914864992, 0.09802547843478299, -0.024592743672618603, -0.09680878245805903, 0.03695114122338155, 0.1584221813761927, 0.0567993226764613, -0.007783348328059781, 0.0023243977612495215, -0.24833154607267083, 0.08656441459947233, 0.07529076629478042, 0.11274801238771782, 0.009826903663616885, -0.21578490050401858, 0.029791279593439288, 0.02260286990751114, 0.08497207067902897, -0.36221665243556406, 0.10395830263116738, 0.04720806491038278, 0.0306570870169622, 0.07202010885962754, -0.2970832322593879, 0.05459798561482855, 0.03089926574912477, 0.11962609939255191, -0.18746907697827264, 0.13182940103245694, 0.060005133634369164, 0.04321100599429057, 0.0897698437111948, -0.17789239663729622, 0.16503240064402075, 0.08417514828530742, 0.05327740917766454, 0.10161052504196755, -0.5218571400901266, 0.03141097382209737, 0.17654534884641296, 0.07109939086056591, 0.033659050640981625, 0.13231195765281642, -0.41484224960883753, 0.013173418407802766, 0.18905642832719868, 0.08614793453501919, 0.11835028344010975, 0.13137985462283933, -0.016484610960276015, -0.21672440861663228, 0.22885747257665554, -0.10584583755253266, 0.08620512776239624, 0.0652162546616284, 0.14518782307999795, 0.13002468379925033, -0.2841153491015078, 0.22348945347611246, -0.09114354598560329, 0.08264069295282177, 0.03753347025334432, 0.1974761773601753, 0.2251162973542438, -0.15162835768732605, 0.08725034273412831, 0.07737463978386097, 0.0546793314919656, 0.23903741283067545, 0.2493601920317472, -0.9579268998032628, 0.16741305003901996, 0.09323380761431421, 0.04652321110975496, 0.21853346416690475, 0.27856268280529983, -0.16147035511325625, 0.1346513745456888, 0.10698892867537117, 0.01513693358800682, 0.1774706618182076, 0.34321198611588605, -0.0945227813759857, 0.2089314815321064, 0.08104878791060759, 0.18403171701865967, 0.3958111101580862, -0.29558887132537937, 0.2291173230575013, 0.013057400043783893, 0.03737146372801951, 0.02433113212704536, 0.040942953241850204, 0.016051687663759303, 0.03187054422267621, 0.0029640032973680406, -0.007722674675589386, 0.013304585484094097, 0.02177060569622866, 0.0381289740905177, 0.0076267453078294085, -0.001882542615129623, 0.03660021547681239, 0.04267002095605698, -0.03634577897853575, 0.013494291386293274, 0.04735519260565965, 0.059609332283440675, 0.02175259685180132, -0.007712439605365152, 0.03559186314237601, 0.03722329015750399, 0.019109144256104504, 0.04482846191206839, 0.01787674666242816, 0.013057543728428815, -0.0108887389010859, 0.055397702916650325, 0.05959980545212032, 0.03814410001231594, 0.028580347799898563, 0.02940557851314421, 0.060220249025983996, 0.026898982596260713, -0.0008491809887510665, 0.0019206894895106483, 0.05201986645907302, 0.058078326661315606, 0.007335417085049373, 0.06358626896774196, 0.032999850925819564, 0.049863154430838934, 0.05172175678965867, 0.020335772510872315, -0.03275631789767349, 0.05248562589963408, 0.06664322010257342, 0.017620755238962074, -0.006120814467506909, -0.04755345753978201, 0.042796268656579706, -0.13864141153666465, 0.0411968109322375, 0.05297928944782069, 0.0032027741459445024, -0.0021714473870729184, 0.044938816047560774, -0.11400532653148082, 0.044689973959106136, 0.054835996661308055, 0.0046993357713335186, -0.08862958163584798, 0.05650340944278794, -0.012127933879809451, 0.03873201956504509, 0.04005399074586799, -0.12457111774146076, -0.001588968318125576, -0.0025281901197995614, 0.04403442956219011, 0.025020891256250535, 0.0799395905706705, 0.043598731103540435, -0.06735779037510405, 0.019733297134846704, -0.003169449785639228, 0.024552089626320925, 0.05333447875337747, 0.040135555148387625, 0.019686892235358484, 0.0017081115903934923, -0.06600170380900207, 0.013371506459887451, 0.06427778061773531, 0.0760479303104155, -0.013933979796083873, -0.007881925215420997, 0.03226639141562891, 0.04332074136698085, 0.06996805442067057, 0.05445703253690702, 0.03355188289292142, -0.020738739044765385, -0.025929681774930435, 0.06572623006725246, 0.08104101153798936, 0.06527940269654589, 0.07617310574264637, -0.0016082986833766773, 0.059873764695370014, 0.09287544357115966, 0.03032285897222144, 0.05693930962579938, 0.22928592646699147, -1.0069601083856794, 0.5712673781614421, 0.75469410620627, 0.2938951594524724, -0.861256724974657, 0.44467609460041846, 0.8089178619469086, 0.4289408908564586, -0.9309819087826372, 0.492061836989798, 1.1435290002055718, 0.8595939887582952, -1.036578969411483, 0.3799840080771718, 0.836452392150436, 0.819220192558215, -0.7575945213434485, 0.39990973577703665, 0.8557951371432629, 0.9091845261135988, -0.49329484948216407, 0.3366381835957261, 0.8790769150951576, -0.640676138867411, 0.5964689537679302, 0.9570546283303178, -0.3711405217382263, 0.32049325269869444, 0.8968718199790096, -0.7755128807303229, 0.44732680278444215, -0.22764625020459672, 0.8963900193572516, 0.9526101184851684, -0.08270711104771217, 0.2558839088204668, 0.9285113505521342, -0.7379885386568785, 0.41322855618901705, -0.22516718285967105, 0.9360541577733569, 0.24065934598090977, 0.9096387504042139, -0.9172770266783825, 0.25203636773688776, -0.18589663953421515, 0.9295540465471075, 0.9652470285717584, 0.3332800673881238, 0.14471375970435946, 0.9319733001684902, -0.9285973573966506, 0.19911295475014268, -0.2253189228894997, 0.931961017709, 0.2354255065061081, 0.9117473636121401, -0.9569261190849616, 0.05995863318518922, -0.2889045074575496, 0.9178604572700467, 0.7331363506735264, 0.718817586196304, 0.1815873610734043, 0.9034803653995013, -0.9144097035484778, -0.03641472471325238, -0.3263786201647654, 0.8919832594596774, 0.6069678800712693, 0.6999816695695548, 0.23463603313771791, 0.9324556554765954, -0.8026783434932715, -0.26186347252480974, -0.5473040176161243, 0.7802173819036111, -0.3937769650061678, 0.7563069798119487, 0.39018165179949293, 0.951768882513349, 0.19539301838724038, 0.9260043611137615, -0.8996939856210328, -0.3343014598476462, -0.5438092638032996, 0.78478246817385, -0.5186796561752327, 0.8024144835561727, 0.24902323603423127, 0.9161515433310885, 0.21089091444181812, 0.9368565049721019, -0.7980393393878066, -0.5324753273937599, -0.5961534686327256, 0.7663115947701238, -0.6893533644656246, 0.6526232379797322, 0.01819210604919684, 1.0187758465206977, -0.5169809579030731, -0.826423160097913, 0.2404794885096374, 0.928504412285588, -0.7187944773832196, -0.6230189324412464, -0.6070450542999236, 0.753942616121833, -0.752768159446818, 0.5844687617167018, -0.0728114222238594, 0.9673352464582473, -0.4534201536922752, -0.8619383618354158, 0.24828300389350924, 0.8451264763300647, -0.3243715800918038, -0.8548982767781169, -0.5845938386116954, 0.7629606656558681, -0.854890895993919, -0.3657630147399848, -1.042165107772652, -0.3187180586307996, 0.35023373688521725, 0.8375380648018983, 0.19963141199170178, -0.9343290208981365, -0.5694286710633558, 0.7898679449094504, 0.04803892347302766, -0.9631995342852346, -0.8602291207209061, 0.47265044399402667, 0.6194079740898284, 1.0631426236004407, 0.7125684697243393, -0.6297990998622047, -0.5262486985613433, 0.802698082638388, 0.6928792710136927, -0.6596715642020232, -0.09390028572241614, 0.9678321576529396, 0.7664282610171284, 1.009040705873495, 0.8629832147774739, -0.2564756251381323, -0.523997509740843, 0.8237617567289012, 0.8848914249989669, -0.3139063772441936, 0.23603700315770273, 0.8996383532764908, 0.9219972468809036, -0.10959613516426085, -0.45483468994157816, 0.8482034654420014, 0.9069750022588081, -0.20464061586356244, 0.4698560303011131, 0.8548330914720841, -0.044600284234313885, 0.9462312438419297, -0.07474722908683319, 0.9425474991485719, -0.04516548946946358, 0.9338725649131636, 0.056884279333648785, 0.9802875622527281, -0.038360251423975276, 0.947548435255813, 0.08002719453225092, 1.4482978012854502, -0.07105579619699881, 1.2703346739235517, 0.042003490341249845, 0.9499516005566803, -0.03477847181308037, 0.9136182769784348, -0.06334452174138976, 0.9399430364509839, -0.06666738820643013, 0.9430722279537006, -0.04609688509168695, 0.9397377807856457, -0.03422805776557725, 0.937078350482144, -0.045809573970890964, 0.9431128893659292, 0.015247317543473023, 0.9597026189052669, 0.03582689822867015, 0.916531285953787, 0.033198527188296044, 0.9253161626521946, -0.05612045434961069, 0.9250008797504263, 0.002431860949764339, 0.9472212813114472, -0.01398813717212549, 0.9206831724576697, 0.030255613667947955, 0.9489530777941105, -0.05266358015497597, 0.9396988746620107, 0.03147680769868421, 0.9422349968935358, -0.008715086168331084, 0.9429030462854604, -0.05370567488595679, 0.9417178736329631, 0.01966426891925546, 0.9414814041524731, 0.005607660393729634, 0.9439064438732025, 0.02167493924904053, 0.9478754820713327, -0.05830968217191396, 0.9396494540643752, -0.04002761038732679, 0.95190053851357, 0.02528797803984323, 0.9543280607081489, -0.03459212874529762, 0.9433782200009281, 0.033258807562186854, 0.9569757063988821, -0.026121304299506442, 0.9578530650149901, 0.0258461865153039, 0.9583551686386873, -0.053340819624953136, 0.9104407295500058, -0.037904677349888546, 0.9474474154736384, -0.01953986810469635, 0.8939892802462388, 0.019507975981269802, 0.9393435543532019, 0.010672577854574986, 0.886820214229917, -0.03997091830897902, 0.9436103949701957, -0.010500045955875899, 0.9436633674075146, -0.05148435634745421, 0.9522974257412768, 0.016629592842047215, 0.94907847634464, 0.05244992097765912, 0.9493220282275651, -0.014157212438132023, 0.9475599996455341, -0.005960676029667083, 0.9504896481798397, -0.08801164887295601, 0.9458930985399897, 0.012246024582715795, 0.953902895290199, 0.05960318464373873, 0.949315459862359, -0.02280353482552486, 0.9429639460601614, -0.5869456363403003, 0.7564081575165513, 0.034021090703852265, 0.9492991487845215, -0.051727270957819746, 0.9423110683404277, 0.013269739463408082, 0.9502786398370566, 0.04800635235855016, 0.9431501008949904, -0.027543871027221105, 0.9559840946834072, -0.23445225565937203, 0.918762782819268, -0.013909096605307685, 0.8914953467767375, -0.07274708527965497, 0.9349132506315386, 0.010267087861764024, 0.9419209340413536, -0.01852669331619129, 0.9530801376928251, 0.28261124164286694, 1.2427114678968603, 0.027423726233106474, 0.927900952959669, 0.05381983629401269, 0.942000515534987, -0.0017770010260220028, 0.9543467589354344, 0.008813231739710535, 0.9535317066735868, 0.14531173770177577, 0.9514500279542246, 0.04835275222575144, 0.8700789945426631, 0.16782811969320363, 0.9326185964924131, -0.003810300132611395, 0.9444533649796364, 0.012321985752411004, 0.9519553746874264, -0.12075506012976639, 0.9486387653089182, 0.06581529477291923, 1.0631029616742493, 0.18761723241786304, 0.9392317666897301, -0.0093884306414966, 0.9560950362912968, -0.01459479435439926, 0.957267318487802, 0.04353481333174155, 0.9515255677249309, 0.16470588502749572, 0.9433635494276872, -0.009254783070661706, 0.9432821229937126, -0.011829308663366797, 0.9538713883436669, -0.024142900385089074, 0.9210216252260249, -0.008609940061830315, -0.05210812875241912, 0.030930864307888536, 0.002642423346309614, 0.0334885304748988, -0.1083662244624674, -0.03300391598567352, -0.03772146039677258, 0.0343411549537251, 0.08734328452022828, 0.0033973523183382743, 0.04553046083737433, 0.012482284609238297, 0.016698408522172938, 0.009037633571012635, 0.020360158896546215, -0.02770928718326479, -0.05578300647528499, -0.004876214228079984, 0.1397084910999681, -0.0409577991078701, -0.03358399545808372, 0.03735991083046108, 0.08638920395384807, 0.06209822104081211, 0.0064312158537089634, 0.00843377025032564, 0.07098347989672463]
Final objective value: -17639.809910195887
)Now let's evaluate our fits by plotting the residuals
using CairoMakie
res = residuals(post, xopt)
fig = Figure(; size = (800, 600))
plotfields!(fig[1, 1], res[1], uvdist, x -> Comrade.measurement(x)[1, 1] / noise(x)[1, 1], axis_kwargs = (; ylabel = "RR Residual"))
plotfields!(fig[2, 1], res[1], uvdist, x -> Comrade.measurement(x)[2, 1] / noise(x)[2, 1], axis_kwargs = (; ylabel = "LR Residual"))
plotfields!(fig[1, 2], res[1], uvdist, x -> Comrade.measurement(x)[1, 2] / noise(x)[1, 2], axis_kwargs = (; ylabel = "RL Residual"))
plotfields!(fig[2, 2], res[1], uvdist, x -> Comrade.measurement(x)[2, 2] / noise(x)[2, 2], axis_kwargs = (; ylabel = "LL Residual"))
fig |> DisplayAs.PNG |> DisplayAs.Text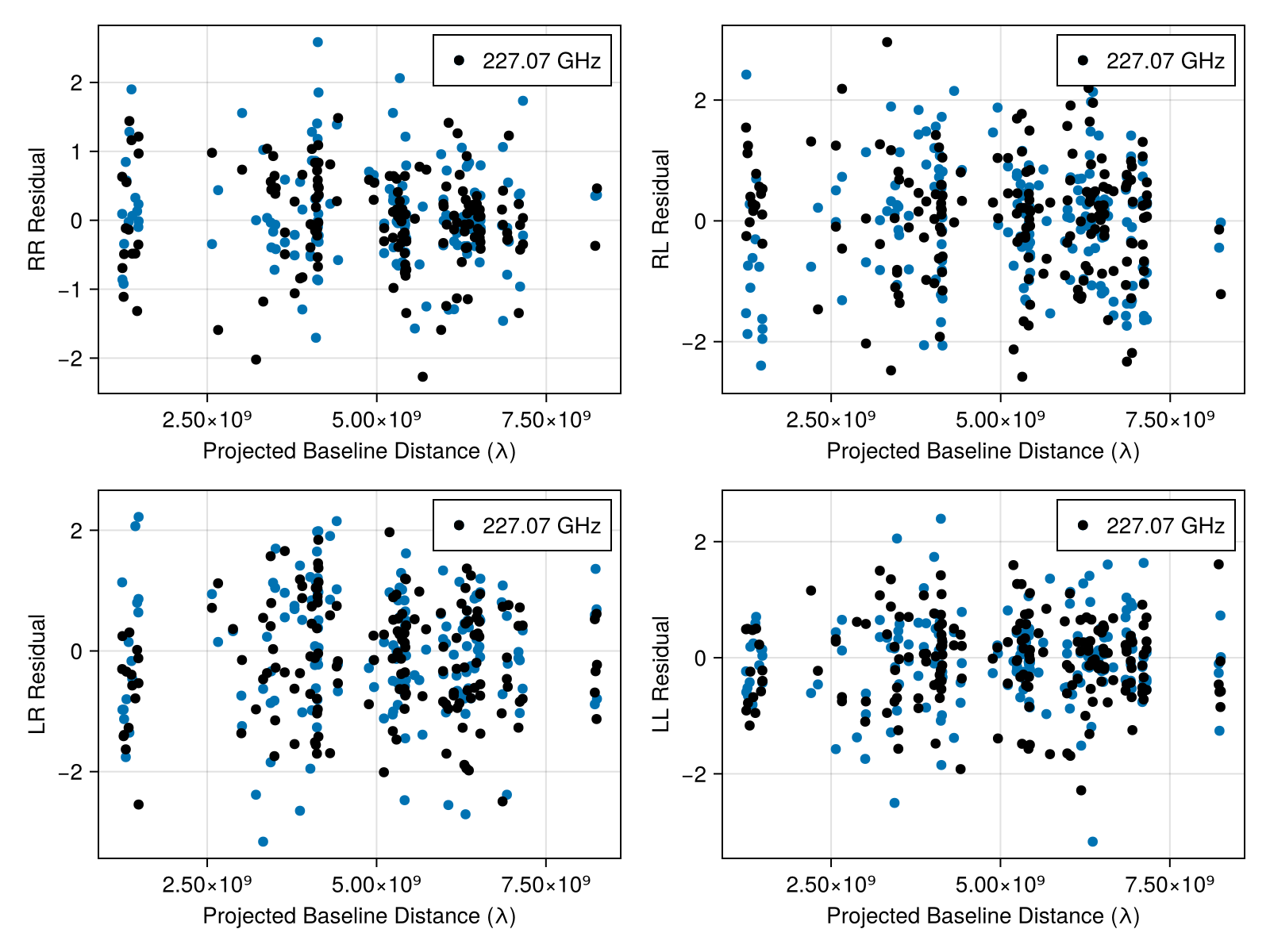
These look reasonable, although there may be some minor overfitting. Let's plot the results
gpl = refinespatial(grid, 2)
img = intensitymap(skymodel(post, xopt), gpl)
fig = imageviz(
img, adjust_length = true, colormap = :cmr_gothic, pcolormap = :rainbow1,
pcolorrange = (0.0, 0.2), plot_total = false
);
fig |> DisplayAs.PNG |> DisplayAs.Text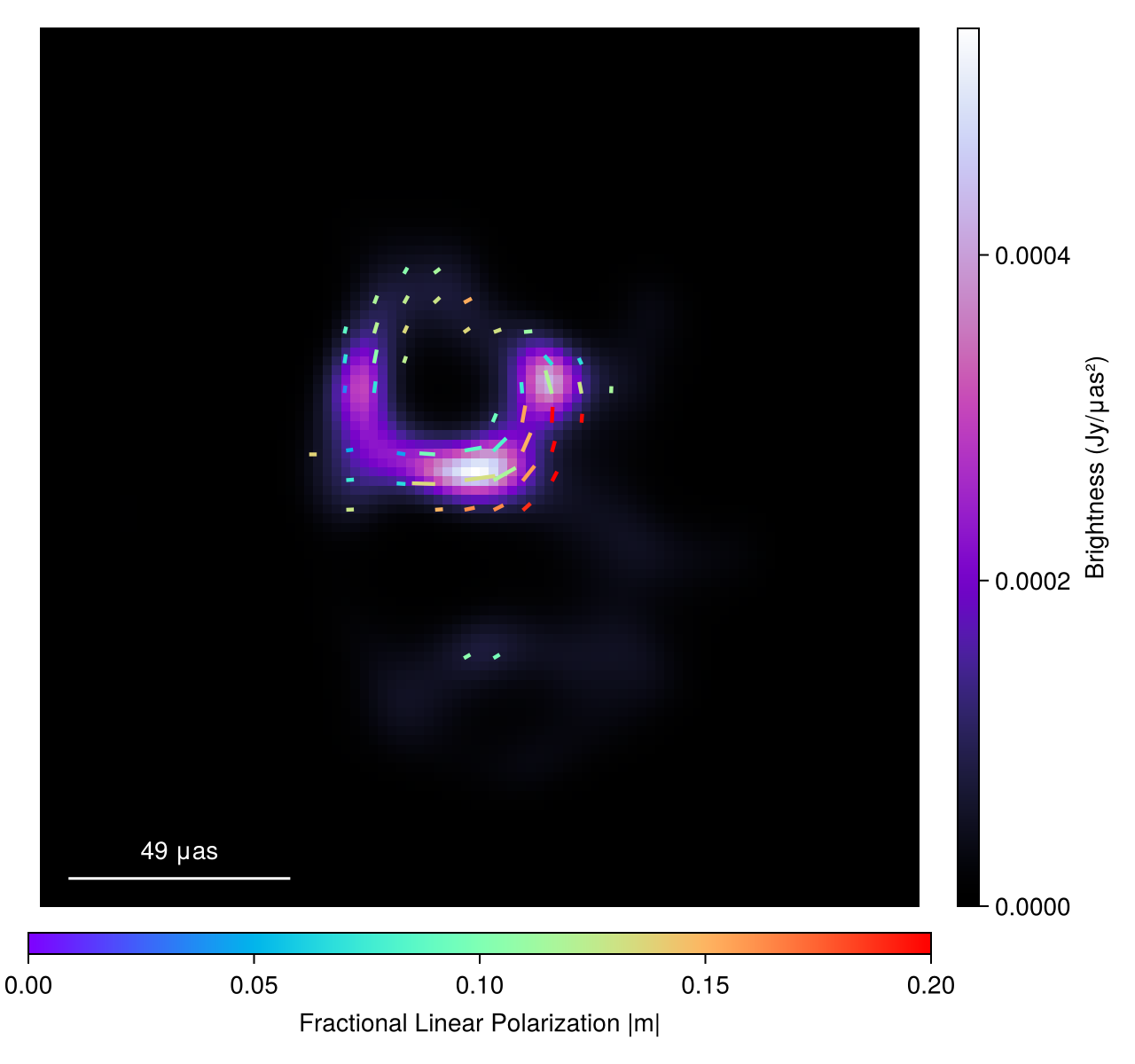
Note
The image looks a little noisy. This is an artifact of the MAP image. To get a publication quality image we recommend sampling from the posterior and averaging the samples. The results will be essentially identical to the results from EHTC VII.
We can also analyze the instrument model. For example, we can look at the gain ratios and products. To grab the ratios and products we can use the caltable function which will return analyze the gprat array and convert it to a uniform table. We can then plot the gain phases and amplitudes.
gphase_ratio = caltable(xopt.instrument.gprat)
gamp_ratio = caltable(exp.(xopt.instrument.lgrat))─────────┬───────────────────┬─────────────────────────────────────────────────────────────
Ti │ Fr │ AA AP AZ JC LM PV SM
─────────┼───────────────────┼─────────────────────────────────────────────────────────────
0.58 hr │ 227.070703125 GHz │ 1.013 missing missing missing missing 1.038 missing
0.73 hr │ 227.070703125 GHz │ 1.025 1.042 missing missing missing 1.016 missing
0.88 hr │ 227.070703125 GHz │ 1.032 1.003 missing missing missing 0.992 missing
1.3 hr │ 227.070703125 GHz │ 1.013 1.022 missing missing missing 1.039 missing
1.45 hr │ 227.070703125 GHz │ 1.008 0.998 missing missing missing 1.037 missing
1.89 hr │ 227.070703125 GHz │ 1.044 0.964 missing missing missing 1.014 missing
2.04 hr │ 227.070703125 GHz │ 1.048 1.061 1.022 missing missing 0.992 missing
2.46 hr │ 227.070703125 GHz │ 1.036 1.038 1.019 missing 1.046 1.018 missing
2.61 hr │ 227.070703125 GHz │ 1.013 0.989 1.057 missing 1.061 missing missing
3.06 hr │ 227.070703125 GHz │ 1.039 1.029 1.03 missing 1.062 1.027 missing
3.21 hr │ 227.070703125 GHz │ 0.999 1.002 1.053 missing 1.06 missing missing
3.62 hr │ 227.070703125 GHz │ 1.007 1.066 1.034 missing 1.051 1.053 missing
3.78 hr │ 227.070703125 GHz │ 1.021 0.968 1.054 missing 1.069 1.018 missing
4.23 hr │ 227.070703125 GHz │ 0.994 0.954 1.044 0.871 1.042 1.054 missing
4.37 hr │ 227.070703125 GHz │ 1.003 0.998 1.046 0.892 1.046 1.056 missing
4.79 hr │ 227.070703125 GHz │ 1.005 0.915 1.058 0.988 1.039 1.041 0.883
4.95 hr │ 227.070703125 GHz │ 0.998 0.997 1.045 1.025 1.083 1.045 0.935
5.63 hr │ 227.070703125 GHz │ 1.02 0.997 1.025 1.055 1.041 missing 1.02
6.15 hr │ 227.070703125 GHz │ 1.002 0.936 1.013 1.066 1.079 missing 0.986
6.72 hr │ 227.070703125 GHz │ 0.992 1.033 1.044 1.072 1.056 missing 1.034
7.23 hr │ 227.070703125 GHz │ 0.979 0.974 1.068 1.084 1.067 missing 1.079
7.44 hr │ 227.070703125 GHz │ 0.998 missing 1.062 1.097 1.031 missing 1.059
─────────┴───────────────────┴─────────────────────────────────────────────────────────────Plotting the phases first, we see large trends in the righ circular polarization phase. This is expected due to a lack of image centroid and the absense of absolute phase information in VLBI. However, the gain phase difference between the left and right circular polarization is stable and close to zero. This is expected since gain ratios are typically stable over the course of an observation and the constant offset was removed in the EHT calibration process.
gphaseR = caltable(xopt.instrument.gpR)
fig = plotcaltable(gphaseR, gphase_ratio, labels = ["R Phase", "L/R Phase"]);
fig |> DisplayAs.PNG |> DisplayAs.Text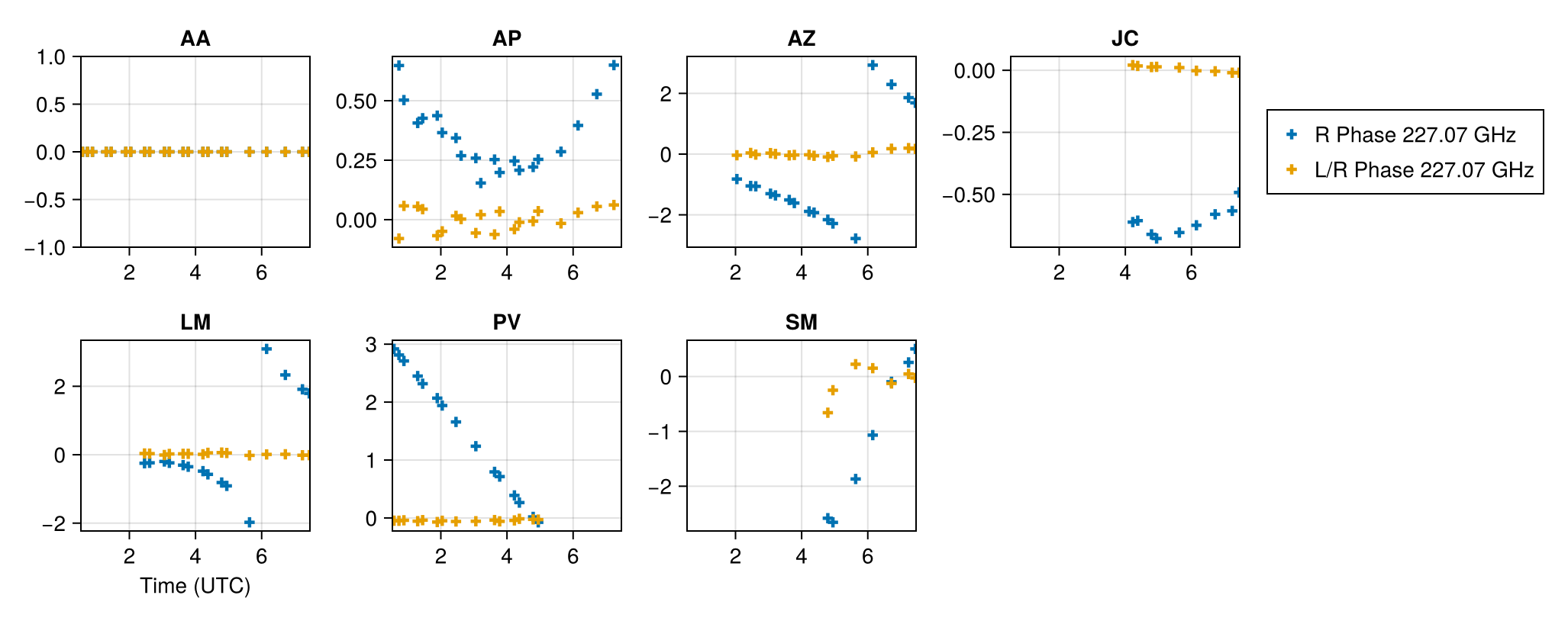
Moving to the amplitudes we see largely stable gain amplitudes on the right circular polarization except for LMT which is known and due to pointing issues during the 2017 observation. Again the gain ratios are stable and close to unity. Typically we expect that apriori calibration should make the gain ratios close to unity.
gampr = caltable(exp.(xopt.instrument.lgR))
fig = plotcaltable(gampr, gamp_ratio, labels = ["R Amp", "L/R Amp"], axis_kwargs = (; limits = (nothing, (0.6, 1.3))));
fig |> DisplayAs.PNG |> DisplayAs.Text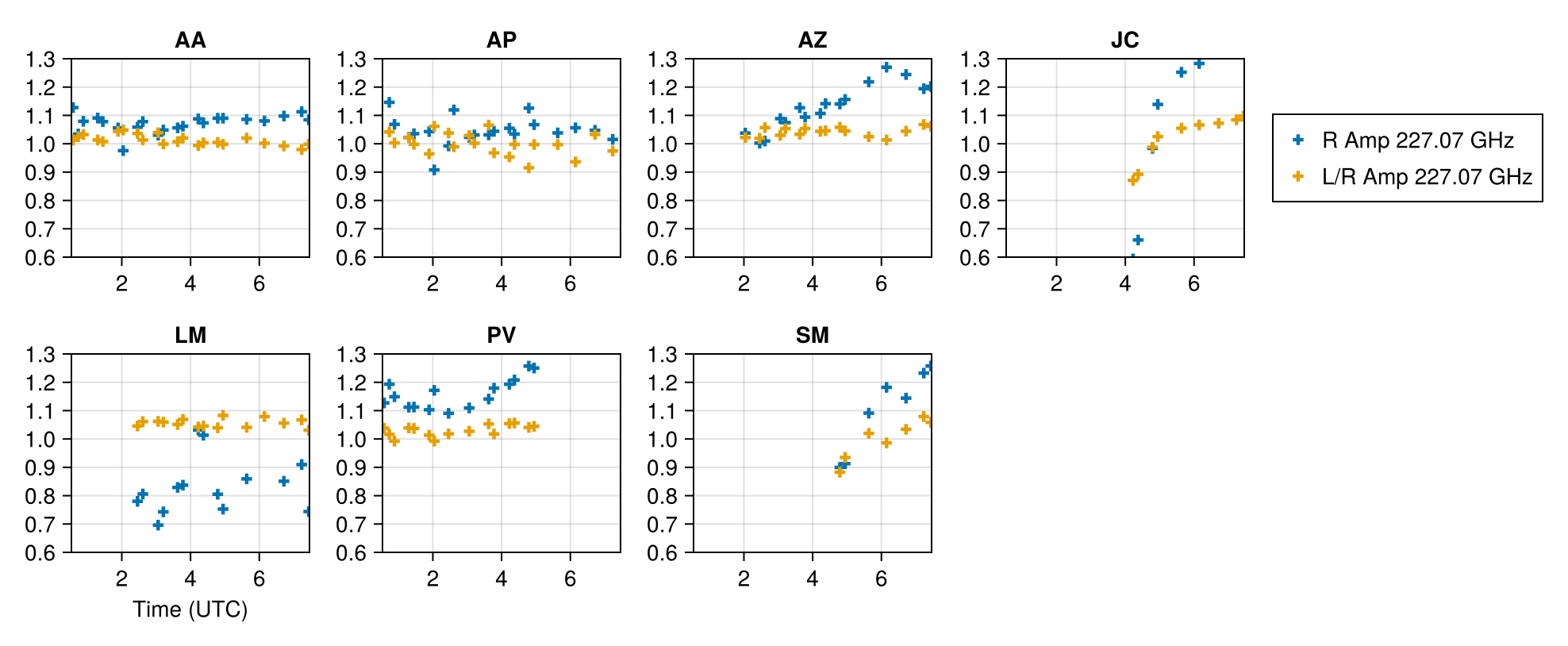
To sample from the posterior, you can then just use the sample function from AdvancedHMC like in the other imaging examples. For example
using AdvancedHMC
chain = sample(rng, post, NUTS(0.8), 2_000, n_adapts = 1000, progress = true, initial_params = xopt)This page was generated using Literate.jl.
Hamaker J.P, Bregman J.D., Sault R.J. (1996) [https://articles.adsabs.harvard.edu/pdf/1996A%26AS..117..137H] ↩︎
Pesce D. (2021) [https://ui.adsabs.harvard.edu/abs/2021AJ....161..178P/abstract] ↩︎