Imaging a Black Hole using only Closure Quantities
In this tutorial, we will create a preliminary reconstruction of the 2017 M87 data on April 6 using closure-only imaging. This tutorial is a general introduction to closure-only imaging in Comrade. For an introduction to simultaneous image and instrument modeling, see Stokes I Simultaneous Image and Instrument Modeling
Introduction to Closure Imaging
The EHT is one of the highest-resolution telescope ever created. Its resolution is equivalent to roughly tracking a hockey puck on the moon when viewing it from the earth. However, the EHT is also a unique interferometer. First, EHT data is incredibly sparse, the array is formed from only eight geographic locations around the planet. Second, the obseving frequency is much higher than traditional VLBI. Lastly, each site in the array is unique. They have different dishes, recievers, feeds, and electronics. Putting this all together implies that many of the common imaging techniques struggle to fit the EHT data and explore the uncertainty in both the image and instrument. One way to deal with some of these uncertainties is to not directly fit the data but instead fit closure quantities, which are independent of many of the instrumental effects that plague the data. The types of closure quantities are briefly described in Introduction to the VLBI Imaging Problem.
In this tutorial, we will do closure-only modeling of M87 to produce a posterior of images of M87.
To get started, we will load Comrade
using ComradePyehtim loads eht-imaging using PythonCall this is necessary to load uvfits files currently.
using PyehtimFor reproducibility we use a stable random number genreator
using StableRNGs
rng = StableRNG(123)StableRNGs.LehmerRNG(state=0x000000000000000000000000000000f7)Load the Data
To download the data visit https://doi.org/10.25739/g85n-f134 To load the eht-imaging obsdata object we do:
obs = ehtim.obsdata.load_uvfits(joinpath(__DIR, "..", "..", "Data", "SR1_M87_2017_096_lo_hops_netcal_StokesI.uvfits"))Python: <ehtim.obsdata.Obsdata object at 0x7f57dfe3ec20>Now we do some minor preprocessing:
Scan average the data since the data have been preprocessed so that the gain phases are coherent.
Add 2% systematic noise to deal with calibration issues such as leakage.
obs = scan_average(obs).add_fractional_noise(0.02)Python: <ehtim.obsdata.Obsdata object at 0x7f5812db5c30>Now, we extract our closure quantities from the EHT data set. We flag now SNR points since the closure likelihood we use is only applicable to high SNR data.
dlcamp, dcphase = extract_table(obs, LogClosureAmplitudes(;snrcut=3), ClosurePhases(;snrcut=3))(EHTObservationTable{Comrade.EHTLogClosureAmplitudeDatum{:I}}
source: M87
mjd: 57849
bandwidth: 1.856e9
sites: [:AA, :AP, :AZ, :JC, :LM, :PV, :SM]
nsamples: 128, EHTObservationTable{Comrade.EHTClosurePhaseDatum{:I}}
source: M87
mjd: 57849
bandwidth: 1.856e9
sites: [:AA, :AP, :AZ, :JC, :LM, :PV, :SM]
nsamples: 152)Note
Fitting low SNR closure data is complicated and requires a more sophisticated likelihood. If low-SNR data is very important we recommend fitting visibilties with a instrumental model.
Build the Model/Posterior
For our model, we will be using an image model that consists of a raster of point sources, convolved with some pulse or kernel to make a ContinuousImage. To define this model we define the standard two argument function sky that defines the sky model we want to fit. The first argument are the model parameters, and are typically a NamedTuple. The second argument defines the metadata for the model that is typically constant. For our model the constant metdata will just by the mean or prior image.
function sky(θ, metadata)
(;fg, c, σimg) = θ
(;mimg) = metadata
# Apply the GMRF fluctuations to the image
rast = apply_fluctuations(CenteredLR(), mimg, σimg.*c.params)
m = ContinuousImage(((1-fg)).*rast, BSplinePulse{3}())
# Force the image centroid to be at the origin
x0, y0 = centroid(m)
# Add a large-scale gaussian to deal with the over-resolved mas flux
g = modify(Gaussian(), Stretch(μas2rad(250.0), μas2rad(250.0)), Renormalize(fg))
return shifted(m, -x0, -y0) + g
endsky (generic function with 1 method)Now, let's set up our image model. The EHT's nominal resolution is 20-25 μas. Additionally, the EHT is not very sensitive to a larger field of views; typically, 60-80 μas is enough to describe the compact flux of M87. Given this, we only need to use a small number of pixels to describe our image.
npix = 32
fovxy = μas2rad(150.0)7.27220521664304e-10To define the image model we need to specify both the grid we will be using and the FT algorithm we will use, in this case the NFFT which is the most efficient.
grid = imagepixels(fovxy, fovxy, npix, npix)RectiGrid(
executor: Serial()
Dimensions:
(↓ X Sampled{Float64} LinRange{Float64}(-3.5224744018114725e-10, 3.5224744018114725e-10, 32) ForwardOrdered Regular Points,
→ Y Sampled{Float64} LinRange{Float64}(-3.5224744018114725e-10, 3.5224744018114725e-10, 32) ForwardOrdered Regular Points)
)Now we need to specify our image prior. For this work we will use a Gaussian Markov Random field prior
using VLBIImagePriors, DistributionsSince we are using a Gaussian Markov random field prior we need to first specify our mean image. For this work we will use a symmetric Gaussian with a FWHM of 50 μas
fwhmfac = 2*sqrt(2*log(2))
mpr = modify(Gaussian(), Stretch(μas2rad(50.0)./fwhmfac))
imgpr = intensitymap(mpr, grid)
skymeta = (;mimg = imgpr./flux(imgpr));Now we can finally form our image prior. For this we use a heirarchical prior where the direct log-ratio image prior is a Gaussian Markov Random Field. The correlation length of the GMRF is a hyperparameter that is fit during imaging. We pass the data to the prior to estimate what the maximumal resolutoin of the array is and prevent the prior from allowing correlation lengths that are much small than the telescope beam size. Note that this GMRF prior has unit variance. For more information on the GMRF prior see the corr_image_prior doc string.
cprior = corr_image_prior(grid, dlcamp)HierarchicalPrior(
map:
ConditionalMarkov(
Random Field: GaussMarkovRandomField
Graph: MarkovRandomFieldGraph{1}(
dims: (32, 32)
)
) hyper prior:
Truncated(Distributions.InverseGamma{Float64}(
invd: Distributions.Gamma{Float64}(α=1.0, θ=0.0407685911951416)
θ: 24.528686684644875
)
; lower=1.0, upper=64.0)
)Putting everything together the total prior is then our image prior, a prior on the standard deviation of the MRF, and a prior on the fractional flux of the Gaussian component.
prior = (c = cprior, σimg = Exponential(0.1), fg=Uniform(0.0, 1.0))(c = HierarchicalPrior(
map:
ConditionalMarkov(
Random Field: GaussMarkovRandomField
Graph: MarkovRandomFieldGraph{1}(
dims: (32, 32)
)
) hyper prior:
Truncated(Distributions.InverseGamma{Float64}(
invd: Distributions.Gamma{Float64}(α=1.0, θ=0.0407685911951416)
θ: 24.528686684644875
)
; lower=1.0, upper=64.0)
)
, σimg = Distributions.Exponential{Float64}(θ=0.1), fg = Distributions.Uniform{Float64}(a=0.0, b=1.0))We can then define our sky model.
skym = SkyModel(sky, prior, grid; metadata=skymeta) SkyModel
with map: sky
on grid: RectiGridSince we are fitting closures we do not need to include an instrument model, since the closure likelihood is approximately independent of gains in the high SNR limit.
using Enzyme
post = VLBIPosterior(skym, dlcamp, dcphase; admode=set_runtime_activity(Enzyme.Reverse))VLBIPosterior
ObservedSkyModel
with map: sky
on grid: FourierDualDomainIdealInstrumentModelData Products: Comrade.EHTLogClosureAmplitudeDatumComrade.EHTClosurePhaseDatumReconstructing the Image
To reconstruct the image we will first use the MAP estimate. This is approach is basically a re-implentation of regularized maximum likelihood (RML) imaging. However, unlike traditional RML imaging we also fit the regularizer hyperparameters, thanks to our interpretation of as our imaging prior as a hierarchical model.
To optimize our posterior Comrade provides the comrade_opt function. To use this functionality a user first needs to import Optimization.jl and the optimizer of choice. In this tutorial we will use Optim.jl's L-BFGS optimizer, which is defined in the sub-package OptimizationOptimJL. We also need to import Enzyme to allow for automatic differentiation.
using Optimization
using OptimizationOptimJL
xopt, sol = comrade_opt(post, LBFGS();
maxiters=1000, initial_params=prior_sample(rng, post))((sky = (c = (params = [-6.452462981261046e-5 0.00032148085346919784 … 0.0002758052859646182 0.00026047832089207693; 0.00010604568097185104 0.00041418480001453336 … 0.0006293163389992453 0.0001610729150537479; … ; 1.7768486235340735e-5 -0.00045492063561373866 … 0.0008359112724586951 0.00033398514971015147; -0.00013776762133456123 -1.7419250556884136e-5 … 0.000734015921382831 0.0001309210305331007], hyperparams = 1.0046571091800072), σimg = 3.0047950690903757, fg = 0.08685775420320072),), retcode: Failure
u: [-6.452462981261046e-5, 0.00010604568097185104, -5.224654945500912e-5, 0.00020200598518007999, 0.0005230825122382017, 0.001123949817931713, 0.0010938847165025407, 0.0008149823905506285, -0.0002763299539510198, -0.0023495119301562063 … -0.00326218245413332, -0.0010627854471878223, -0.00016564655721719093, 0.0006932690238118018, 0.0005201461437528048, 0.00033398514971015147, 0.0001309210305331007, -9.512421172268926, 1.1002093690195363, -2.3526198974841757]
Final objective value: -4.339081213944524
)First we will evaluate our fit by plotting the residuals
using Plots
p = residual(post, xopt)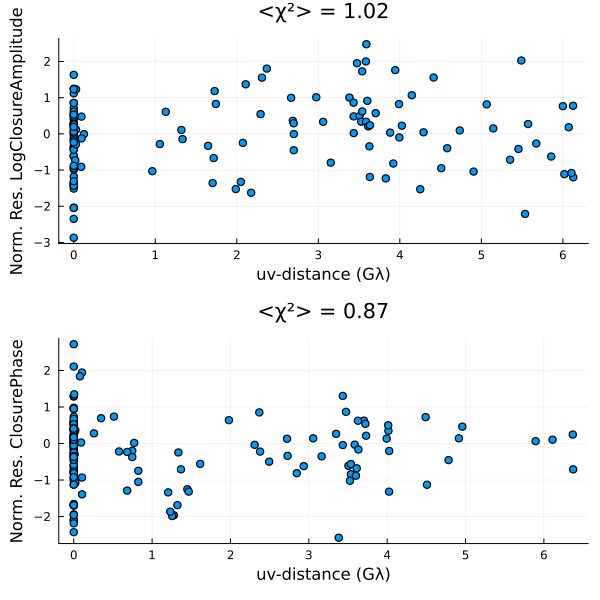
Now let's plot the MAP estimate.
import CairoMakie as CM
g = imagepixels(μas2rad(150.0), μas2rad(150.0), 100, 100)
img = intensitymap(skymodel(post, xopt), g)
fig = imageviz(img, size=(600, 500));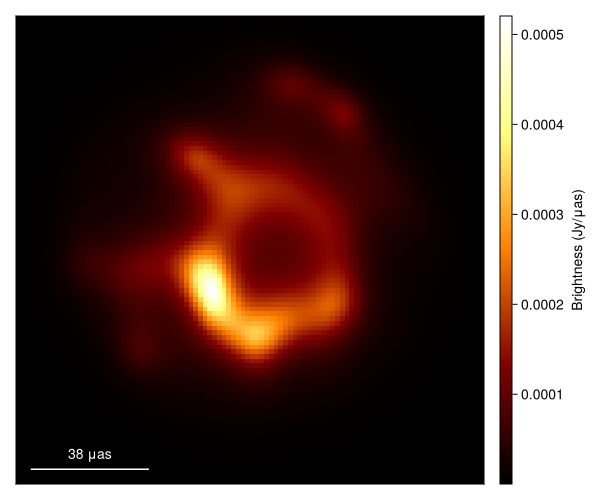
That doesn't look great. This is pretty common for the sparse EHT data. In this case the MAP often drastically overfits the data, producing a image filled with artifacts. In addition, we note that the MAP itself is not invariant to the model parameterization. Namely, if we changed our prior to use a fully centered parameterization we would get a very different image. Fortunately, these issues go away when we sample from the posterior, and construct expectations of the posterior, like the mean image.
To sample from the posterior we will use HMC and more specifically the NUTS algorithm. For information about NUTS see Michael Betancourt's notes.
Note
For our metric we use a diagonal matrix due to easier tuning.
using AdvancedHMC
chain = sample(rng, post, NUTS(0.8), 700; n_adapts=500, progress=false, initial_params=xopt);[ Info: Found initial step size 0.003125Warning
This should be run for longer!
Now that we have our posterior, we can assess which parts of the image are strongly inferred by the data. This is rather unique to Comrade where more traditional imaging algorithms like CLEAN and RML are inherently unable to assess uncertainty in their reconstructions.
To explore our posterior let's first create images from a bunch of draws from the posterior
msamples = skymodel.(Ref(post), chain[501:2:end]);The mean image is then given by
using StatsBase
imgs = intensitymap.(msamples, Ref(g))
mimg = mean(imgs)
simg = std(imgs)
fig = CM.Figure(;resolution=(700, 700));
axs = [CM.Axis(fig[i, j], xreversed=true, aspect=1) for i in 1:2, j in 1:2]
CM.image!(axs[1,1], mimg, colormap=:afmhot); axs[1, 1].title="Mean"
CM.image!(axs[1,2], simg./(max.(mimg, 1e-8)), colorrange=(0.0, 2.0), colormap=:afmhot);axs[1,2].title = "Std"
CM.image!(axs[2,1], imgs[1], colormap=:afmhot);
CM.image!(axs[2,2], imgs[end], colormap=:afmhot);
CM.hidedecorations!.(axs)
fig |> DisplayAs.PNG |> DisplayAs.Text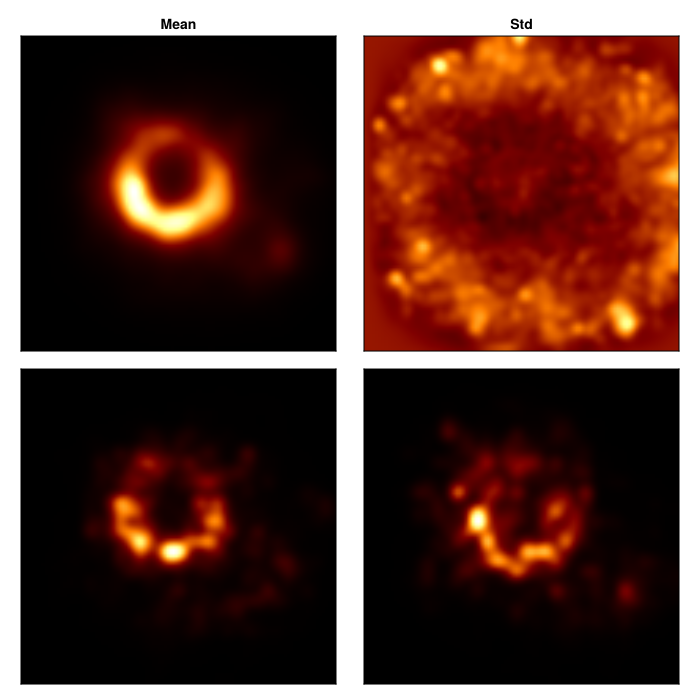
Now let's see whether our residuals look better.
p = Plots.plot(layout=(2,1));
for s in sample(chain[501:end], 10)
residual!(post, s)
end
p |> DisplayAs.PNG |> DisplayAs.Text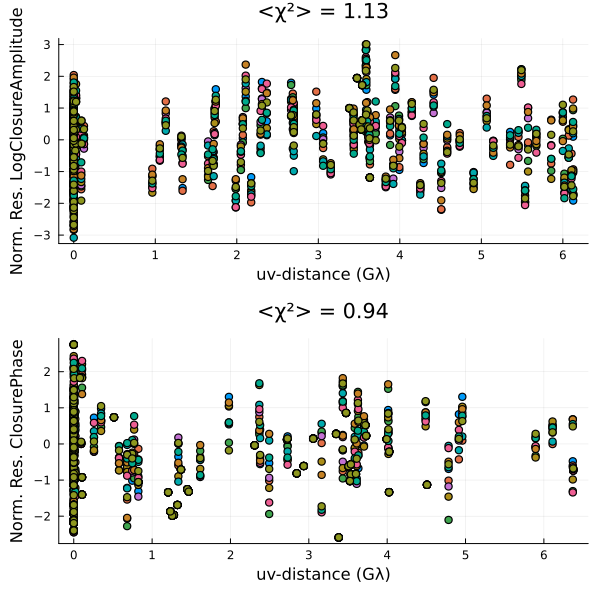
And viola, you have a quick and preliminary image of M87 fitting only closure products. For a publication-level version we would recommend
Running the chain longer and multiple times to properly assess things like ESS and R̂ (see Geometric Modeling of EHT Data)
Fitting gains. Typically gain amplitudes are good to 10-20% for the EHT not the infinite uncertainty closures implicitly assume
This page was generated using Literate.jl.